Abstract
Two novel diarylcyclopentenones daturamycin A and B (1 and 2), and one new p-terphenyl daturamycin C (3), along with three known congeners (4–6), were isolated from a rhizosphere soil-derived Streptomyces sp. KIB-H1544. The structures of new compounds were elucidated via a joint use of spectroscopic analyses and single-crystal X-ray diffractions. Compounds 1 and 2 belong to a rare class of tricyclic 6/5/6 diarylcyclopentenones, and compounds 3–6 possess a C-18 tricyclic aromatic skeleton. The biosynthetic gene cluster of daturamycins was identified through gene knockout and biochemical characterization experiments and the biosynthetic pathway of daturamycins was proposed.
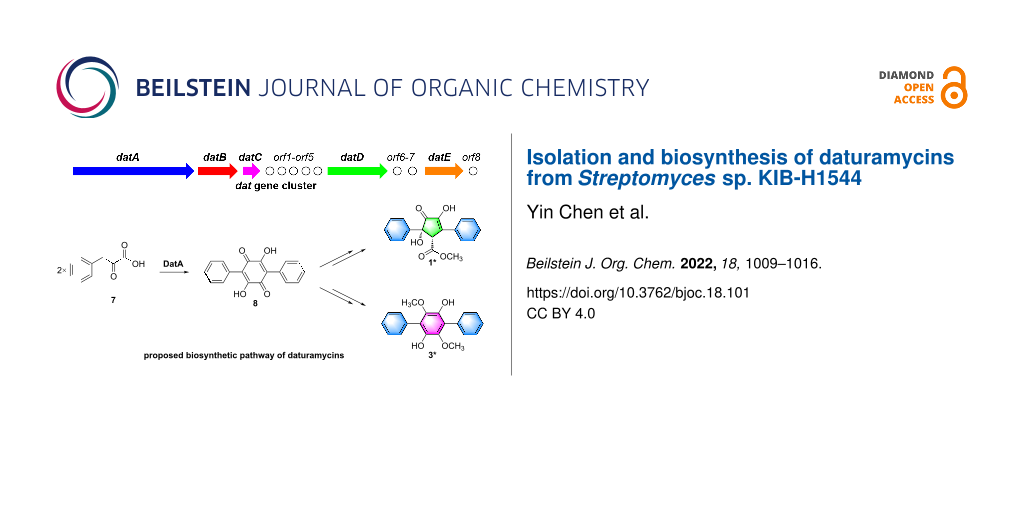
Graphical Abstract
Introduction
Natural products containing a terphenyl skeleton exhibit a large number of structural diversity due to the differences of the center ring and the connection among rings. Structurally, most natural terphenyls are p-terphenyl derivatives consisting of a C-18 tricyclic or polycyclic C-18 aromatic skeleton. Diarylcyclopentenones, which possess a rare class of a tricyclic 6/5/6 system, could also belong to p-terphenyl derivatives from a biosynthetic perspective. To date, more than 230 natural products containing p-terphenyl have been unearthed in nature, among them many p-terphenyls were isolated from fungi, only a few were discovered from Streptomyces species [1-4]. Meanwhile, these types of compounds exhibit a broad spectrum of bioactivities, including antitumor [5,6], antibacterial [7,8], antioxidant [9-12], immunosuppressive [13], and antithrombotic activities [14].
The biosynthesis of the p-terphenyls has been studied since the 1960s [15,16]. Stable-isotope labeling experiments confirmed that ʟ-tyrosine or ʟ-phenylalanine are involved in the biosynthesis of p-terphenyl as metabolic origin. The precursors undergoing deamination are converted to the corresponding α-keto acid, then a quinone intermediate arises by condensation between two molecules of α-keto acid. Structurally diverse p-terphenyls are formed from these key intermediates by several tailoring reactions such as cyclization, tautomerization, methylation, and glycosylation. A previous study has shown that the formation of 2,5-diarylcyclopentenone proceeds via the terphenylquinone atromentin (Figure 1), followed by oxidative ring contraction [17]. However, the details of cyclopentenone formation involving ring contraction have remained unclear.
Figure 1: Structures of compounds 1–6, atromentin, and echoside C.
Figure 1: Structures of compounds 1–6, atromentin, and echoside C.
In this study, two novel diarylcyclopentenones daturamycins A amd B (1 and 2), one new p-terphenyl daturamycin C (3), and three known p-terphenyl derivatives (4–6) (Figure 1) were isolated from the fermentation extract of Streptomyces sp. KIB-H1544. Structurally, daturamycins A and B are uncommon diarylcyclopentenone compounds that consist of a tricyclic 6/5/6 system instead of a C-18 tricyclic skeleton in other compounds. To explore the biosynthesis of daturamycins in S. sp. KIB-H1544, especially the formation mechanism of the tricyclic 6/5/6 scaffold, the biosynthetic gene cluster of daturamycins was found and confirmed by gene knockout experiments. We also characterized the deduced peptide synthetase DatA, which catalyzes the Claisen–Dieckmann condensation of phenylpyruvic acid (7) to generate the key intermediate polyporic acid (8). Finally, we proposed a biosynthetic pathway for daturamycins.
Results and Discussion
Daturamycin A (1), a yellow powder, possessed a molecular formula of C19H16O5 with 12 degrees of unsaturation based on the HRMS–ESI data (m/z 347.0893 [M + Na]+, calcd for C19H16O5Na+, 347.0890) (Figure S1, Supporting Information File 1). Comprehensive analysis of the 1H and 13C NMR data (Table 1, Figures S3 and S4, Supporting Information File 1) and HSQC data (Figure S5, Supporting Information File 1) demonstrated the presence of one methyl, eleven methines (including ten aromatic ones), and seven quaternary carbons (including two carbonyl carbons). The structural information of 1 was further defined according to HMBC data (Figure S6, Supporting Information File 1). The key HMBC signals (H-2’ to C-1’, C-4’, C-6’; H-3’ to C-1’, C-5’; H-4’ to C-2’, C-6’ ; H-5’ to C-1’, C-3’; H-2’’ to C-1’’, C-4’’, C-6’’; H-3’’ to C-1’’, C-5’’; H-4’’ to C-2’’, C-6’’, and H-5’’ to C-1’’, C-3’’) of compound 1 indicated the presence of two mono-substituted aromatic rings. The cross-peaks from H-4 to C-1, C-2, C-3, C-5, and C-6, from 6-OCH3 to C-6 established a central five-membered carbon ring. Meanwhile, the correlations from H-4 to C-1’, C-1’’, from H-2’ and H-6’ to C-5, and from H-2’’ and H-6’’ to C-3 showed that two benzene rings were conjugated to the central ring at the C-3 and C-5 positions (Figure 2). The NMR data indicated that compound 1 shares a similar skeleton to the known compound (±)-tylopilusin B [18]. The additional signals suggested that three methines (δC/δH 59.2/4.49, CH-4; 129.0/7.27, CH-4’; 130.5/7.38, CH-4’’) in compound 1 were hydroxylated in (±)-tylopilusin B. Furthermore, the absolute configuration of C-4 and C-5 in compound 1 was also confirmed as 4R and 5S by X-ray crystallography (Figure 2).
Table 1: 1H (600 MHz) and 13C (150 MHz) NMR data of compounds 1 and 2.
| 1 (in CD3OD) | 2 (in DMSO-d6) | |||
| position | δC | δH, mult. (J in Hz) | δC | δH, mult. (J in Hz) |
| 1 | 201.2, C | 204.6, C | ||
| 2 | 151.4, C | 140.8, C | ||
| 3 | 135.1, C | 157.7, CH | 7.98, d (1.8) | |
| 4 | 59.2, CH | 4.49, s | 77.5, CH | 4.84, d (4.3) |
| 5 | 79.4, C | 85.5, C | ||
| 6 | 172.8, C | |||
| 1’ | 143.5, C | 139.6, C | ||
| 2’/6’ | 126.2, CH | 7.44, d (7.7) | 126.9, CH | 7.28, m |
| 3’/5’ | 129.5, CH | 7.33, t (7.5) | 127.3, CH | 7.28, m |
| 4’ | 129.0, CH | 7.27, t (7.3) | 126.7, CH | 7.21, t (6.9) |
| 1’’ | 134.7, C | 130.7, C | ||
| 2’’/6’’ | 128.7, CH | 7.95, d (7.7) | 127.1, CH | 7.82, d (7.3) |
| 3’’/5’’ | 129.8, CH | 7.44, t (7.5) | 128.6, CH | 7.45, t (7.3) |
| 4’’ | 130.5, CH | 7.38, t (7.3) | 129.0, CH | 7.41, t (7.3) |
| 6-OCH3 | 52.9, CH3 | 3.71, s | ||
| 4-OH | 5.60, d (6.3) | |||
| 5-OH | 6.29, s | |||
![[1860-5397-18-101-2]](/bjoc/content/figures/1860-5397-18-101-2.png?scale=2.0&max-width=1024&background=FFFFFF)
Figure 2: (A) Key 2D NMR correlations of compounds 1 and 2. (B) X-ray crystal structure of compounds 1 and 3.
Figure 2: (A) Key 2D NMR correlations of compounds 1 and 2. (B) X-ray crystal structure of compounds 1 and 3.
(±)-Daturamycin B (2) was isolated as a white powder, and its molecular formula was determined as C17H14O3 by HRMS–ESI data (m/z 289.0833 [M + Na]+, calcd for C19H16O5Na+, 289.0835) (Figure S7, Supporting Information File 1), and the unsaturation was 11 degree. It could be deduced that compound 2 also possesses a similar core to compound 1, based on the molecular formula and NMR data (Table 1, Figures S8–S13, Supporting Information File 1). This could be confirmed by the key COSY correlation of H-3/H-4 and the HMBC correlations (H-3 to C-1, C-4, C-1’’; H-4 to C-1, C-2, C-1’; 4-OH to C-3, C-5; 5-OH to C-1, C-4, C-1’; H-2’ to C-5; H-3’ to C-1’; H-4’ to C-2’, C-6’; H-2’’ to C-2, C-3, C-4’’; H-3’’ to C-1’’; H-4’’ to C-2’’). The NOESY correlation (Figure 2) between H-4 and 5-OH suggested that the relative configurations of C-4 and C-5 were trans. Therefore, the structure of compound 2 has been determined, as shown in Figure 1. However, the absolute configuration of compound 2 remained unsolved.
Daturamycin C (3) was obtained as a brown powder, and the 1H and 13C NMR spectra (Figures S14 and S15, Supporting Information File 1) indicated that compound 3 showed strong similarity with 1,4-diphenyl-2,3,5,6-tetramethoxybenzene [19], with two methyl signals were absent in the NMR data of 3. Furthermore, the structure of 3 could be confirmed, as shown in Figure 1, based on X-ray crystallography (Figure 2).
The known congeners were determined as terferol (4) [20], BTH-II0204-207: D (5) [21], and betulinan A (6) [22] (Figure 1) by comparing their 1H and 13C NMR spectroscopic data (Figures S16–S21, Supporting Information File 1) and specific rotation values with those in the literatures.
Diarylcyclopentenones, characteristic constituents of mushrooms [23], were rarely discovered in Streptomyces species. These components exhibit redox activity and are involved in reducing ferric (Fe3+) in the Fenton-based biological decomposition of lignocellulose [24,25]. The biosynthetic pathway of p-terphenyl was previously identified in Paxillus involutus, and atromentin was supposed to be a metabolic precursor of diarylcyclopentenone [17]. However, the mechanism of cyclopentenone moiety formation has remained unclear. Two pathways may be involved in the biosynthesis of diarylcyclopentenone (Figure 3B): (path a) two molecules of phenylpyruvic acid (7) undergo direct condensation to form a five-membered ring intermediate or (path b) polyporic acid (8) undergoes oxidative ring contraction or conversion to generate the cyclopentenone skeleton.
Figure 3: (A) The biosynthetic gene cluster of daturamycins. (B) Proposed biosynthetic pathway of daturamycins.
Figure 3: (A) The biosynthetic gene cluster of daturamycins. (B) Proposed biosynthetic pathway of daturamycin...
To verify which pathway was responsible for the biosynthesis of daturamycins in S. sp. KIB-H1544, we conducted the following experiments. Firstly, the genome of S. sp. KIB-H1544 was sequenced, and bioinformatic analysis yields the three-genes cluster (NCBI accession number: ON973849) encoded by datA (peptide synthetase), datB (NAD-dependent dehydratase), and datC (limonene-1,2-epoxide hydrolase), which share high sequence similarity with the echoside (Figure 1) biosynthetic gene cluster [26] (EchA, 69.2% identity to DatA; EchB, 78.9% identity to DatB; EchC, 66.9% identity to DatC) (Figure 3A). Additionally, one bilirubin oxidase (DatD) and NADPH-dependent oxidoreductase (DatE) are found downstream of the three-genes cluster (Table S1, Supporting Information File 1). Subsequently, we set out to knockout datA gene by λ-RED-mediated recombination in S. sp. KIB-H1544. Culture and fermentation results of mutant S. sp. KIB-H1544-∆datA indicated that the primary production of compounds 1–6 in the mutant was eliminated (Figure 4A). This result suggested that the datA gene is responsible for the biosynthesis of daturamycins. Lastly, in order to verify the cyclopentenone rings of daturamycins A and B are generated via direct condensation of phenylpyruvic acid (7) catalyzed by DatA, the DatA protein was expressed and purified (Figure 4B). Incubating DatA with substrate 7 and ATP resulted in a new product 8, which was confirmed by HRMS–ESI data (m/z 291.0663 [M − H]−, calcd for ([C18H12O4] − H)−, 291.0663) (Figure S22, Supporting Information File 1). The product was not present in the control reaction with boiled DatA protein (Figure 4B). Furthermore, we could not detect any other intermediates in the reaction mixture through MS analysis (Figure S23, Supporting Information File 1). These results suggested that DatA could not catalyze the formation of the cyclopentenone ring. It has the same function as EchA, which only catalyzes the Claisen–Dieckmann condensation of phenylpyruvic acid (7) to generate polyporic acid (8) [26].
![[1860-5397-18-101-4]](/bjoc/content/figures/1860-5397-18-101-4.png?scale=2.0&max-width=1024&background=FFFFFF)
Figure 4: (A) HPLC analysis of the fermentation extracts of mutant S. sp. KIB-H1544-∆datA. (B) SDS-PAGE analysis of purified DatA (calculated molecular mass 101.8 kDa) and EchA (calculated molecular mass 105.4 kDa) protein. (C) HPLC analysis of in vitro enzyme reactions. Panel I, phenylpyruvic acid with boiled DatA; panel II, phenylpyruvic acid with EchA; panel III, phenylpyruvic acid with DatA.
Figure 4: (A) HPLC analysis of the fermentation extracts of mutant S. sp. KIB-H1544-∆datA. (B) SDS-PAGE analy...
Based on the gene knockout and biochemical characterization experiments, we proposed a possible biosynthetic pathway for daturamycins in S. sp. KIB-H1544 (Figure 3B). First, two molecules of phenylpyruvic acid (7), which is deaminated from ʟ-phenylalanine, are condensed into the six-membered polyporic acid (8) catalyzed by DatA. Then, 8 converts to compounds 3, 4, 5, and 6 through methylation and oxidation. On the other hand, polyporic acid (8) probably undergoes rearrangement, decarboxylation, methylation reaction, and further modification to generate compounds 1 and 2.
Conclusion
S. sp. KIB-H1544 was isolated from the rhizosphere soil of Datura stramonium L. Two novel diarylcyclopentenones daturamycins A and B and one new p-terphenyl daturamycin C, were isolated and identified. The new diarylcyclopentenones possess a rare class of a tricyclic 6/5/6 system. Based on the bioinformatics analysis, the daturamycins biosynthetic gene cluster has been identified and confirmed by the inactivation of the structural gene datA. The following biochemical characterization indicated that the DatA is a polyporic acid synthetase, which helped to propose a possible biosynthetic pathway for daturamycins in S. sp. KIB-H1544 (Figure 3B). Finally, the discovery of daturamycins, identification, and characterization of dat biosynthetic gene cluster allowed us to explore their biosynthesis, especially the mechanism of cyclopentenone formation in the diarylcyclopentenones.
Experimental
General experimental procedures
IR spectra were measured on a Bruker Tensor 27 FTIR spectrometer using KBr disks. NMR spectra were performed with a Bruker AV600 MHz spectrometer with TMS as an internal standard. HRMS–ESI data were obtained through an Agilent 1290 UPLC/6540 Q-TOF mass instrument. Column chromatography (CC) was performed using silica gel (30–400 mesh, Qingdao Marine Chemical Inc., China), Sephadex LH-20 (25–100 µm, Pharmacia Biotech Ltd., Sweden), and MCI gel (75–150 µm, Mitsubishi Chemical Corporation, Tokyo, Japan). Semipreparative HPLC was conducted on a HITACHI Chromaster system equipped with a DAD detector, a YMC-Triart C18 column (250 × 10 mm, i.d. 5 μm), and a flow rate of 3.0 mL/min. LC–MS was performed using an Agilent 1200 series HPLC system coupled to an Agilent q TOF 6540 mass spectrometer with a YMC-Triart C18 column (250 × 4.6 mm, i.d. 5 μm) and a flow rate of 1.0 mL/min.
Extraction and isolation
S. sp. KIB-H1544, isolated from the rhizosphere soil of Datura stramonium L., was 99.9% similar to the 16S rRNA of Streptomyces roseofulvus NBRC15816. The strain was cultured in 20 L M15 medium: 30.0 g/L glucose, 1.0 g/L peptone, 5.0 g/L beef paste, 2.5 g/L CaCO3, 5.0 g/L NaCl, 1.0 mL/L trace elements solution (1.0 g/L FeSO4·7H2O, 0.45 g/L CuSO4·5H2O, 1.0 g/L ZnSO4·7H2O, 0.1 g/L MnSO4·4H2O, 0.1 g/L K2MoO4). After extraction and concentration, the extract was purified by silica gel chromatography and semipreparative HPLC to yield daturamycin A (1) (4.5 mg), daturamycin B (2) (1.8 mg), daturamycin C (3) (8.7 mg), terferol (4) (2.4 mg), BTH-II0204-207: D (5) (2.1 mg), betulinan A (6) (6.2 mg).
Identification of new compounds
Daturamycin A (1): pale yellow solid; IR (KBr) νmax: 3468, 3287, 1731, 1702, 1631, 1398 and 1202 cm−1; UV (MeOH) λmax (log ε): 314 (0.48), 217 (0.28), 202 (0.48); 1H and 13C NMR, see Table 1; HRMS–ESI (m/z): [M + Na]+ calcd for C19H16O5Na+, 347.0890; found, 347.0893.
Daturamycin B (2): white powder; 1H and 13C NMR, see Table 1; HRMS–ESI (m/z): [M + Na]+ calcd for C19H16O5Na+, 289.0835; found, 289.0833.
Daturamycin C (3): brown powder; 1H NMR (600 MHz, CDCl3) δ 7.57 (d, J = 7.2 Hz, 4H), 7.48 (t, J = 7.6 Hz, 4H), 7.39 (t, J = 7.4 Hz, 2H), 5.52 (s, 2H), 3.38 (s, 6H); 13C NMR (150 MHz, CDCl3) δ 140.9, 139.6, 132.8, 130.6, 130.3, 128.3, 128.0, 127.7, 120.7.
X-ray crystal data of compound 1: C19H16O5, M = 324.32, a = 9.8765(14) Å, b = 8.9786(13) Å, c = 18.315(3) Å, α = 90°, β = 92.119(3)°, γ = 90°, V = 1623.0(4) Å3, T = 100(2) K, space group P21/n, Z = 4, μ(Mo Kα) = 0.096 mm−1, 16091 reflections measured, 4037 independent reflections (Rint = 0.0728). The final R1 values were 0.0526 (I > 2σ(I)). The final wR(F2) values were 0.1003 (I > 2σ(I)). The final R1 values were 0.1061 (all data). The final wR(F2) values were 0.1183 (all data). The goodness of fit on F2 was 1.003.
X-ray crystal data of compound 3: C20H18O4, M = 322.34, a = 5.9209(8) Å, b = 20.882(3) Å, c = 7.0856(10) Å, α = 90°, β = 113.939(2)°, γ = 90°, V = 800.70(19) Å3, T = 100(2) K, space group P21/c, Z = 2, μ(Mo Kα) = 0.093 mm−1, 8836 reflections measured, 2398 independent reflections (Rint = 0.0250). The final R1 values were 0.0423 (I > 2σ(I)). The final wR(F2) values were 0.1059 (I > 2σ(I)). The final R1 values were 0.0495 (all data). The final wR(F2) values were 0.1101 (all data). The goodness of fit on F2 was 1.051.
Genomic library construction and screening
The genome of S. sp. KIB-H1544 was sequenced through the Illumina Genome Analyzer (Illumina, San Diego, CA) by BGI (BGI-Shenzhen, China). According to standard procedures, the genomic DNA was digested with MboI, and the 30–42 kb DNA fragments were isolated and ligated to cosmid pSuperCos I. MaxPlax Lambda packaging extracts were used for packaging. About 2,000 E. coli clones were picked and stored in 20 96-well microplates at −80 °C. The positive clone, named pSC-21A4, was verified by PCR. And primers used for screening were listed in Table S3 (Supporting Information File 1).
Gene inactivation
Primers designed for inactivation of datA gene are listed in Table S3 (Supporting Information File 1). The deletion mutant was constructed by λ-RED-mediated PCR targeting mutagenesis method [27]. The positive cosmid pSC-21A4 was transformed into E. coli BW25113/pIJ790 for gene inactivation. Then, the PCR fragment was introduced via electro-transformation into E. coli BW25113/pIJ790-pSC-21A4, in which the target gene datA would be replaced with the cassette. This recombinant cosmid was transformed into E. coli ET12567/pUZ8002, and suffered from intergeneric conjugation with S. sp. KIB-H1544 wild strain. E. coli-Streptomyces conjugation was performed on MS solid medium freshly supplemented with 10 mM MgCl2. Double crossover mutants were selected based on the Kan-Apr and then confirmed by PCR using primers. Finally, the mutant strain S. sp. KIB-H1544-∆datA was generated (Table S2, Supporting Information File 1).
Expression and purification of recombinant DatA and EchA protein
The DNA fragment containing datA was amplified by the primer pair Duet-DatA-F/Duet-DatA-R (Table S3, Supporting Information File 1), then cleaved by BamH I and Sac I, and inserted into the corresponding site of pETDuet-sfp to produce plasmid pETDuet-sfp-datA. As a positive control, the DNA fragment containing echA gene (GenBank: KJ156360.1) was synthesized by GENEWIZ company, plasmid pETDuet-sfp-echA was constructed using the same method as DatA expression vector construction. The recombinant plasmids were then transformed into E. coli BL21 (DE3), respectively. pETDuet-sfp-datA and pETDuet-sfp-echA were grown in Luria Bertani (LB) supplemented with 100 μg/mL ampicillin at 37 °C until OD600 reached 0.6. IPTG (0.2 mM) was added to the final concentration of 0.4 mM and incubated at 16 °C overnight. The cells were centrifuged at 4000 rpm for 25 min and resuscitated with lysis buffer (buffer A: 50 mM Tris-HCl, 300 mM NaCl, 20 mM imidazole, pH 8.0). After ultrasonic cell crushing, the cells were centrifuged at 24,000 rpm for 60 min to remove cell fragments. The supernatant was filtered through 0.22 μm of the filter membrane and then loaded into the nickel column of the rebalanced lysate (HisTraqTM FF, GE Healthcare). The eluent was removed with buffer B (50 mM Tris-HCl, 300 mM NaCl, 250 mM imidazole, pH 8.0) at a flow rate of 2 mL/min, starting from the fourth tube of the collection tube and ending at the sixth tube. The outgoing protein was collected and poured into a precooled filter column (Ultracel series 10 KDa; GE Healthcare). Then, the collected DatA (or EchA) protein was concentrated and buffer-exchanged into storage buffer (50 mM NaH2PO4, 100 mM NaCl, pH 8.0, containing 10% glycerin). The proteins were tested by SDS-PAGE, frozen using liquid nitrogen, and kept at −80 °C.
In vitro reactions
DatA enzymatic activity was tested in a 100 μL reaction mixture containing 100 mM Tris-HCl (pH 7.5), 10 mM MgCl2, 300 mM NaCl, 5 mM ATP, 0.5 mM phenylpyruvic acid and 5 μM DatA (or EchA), at 30 °C for 2 h. The reaction mixture was quenched by adding 200 μL methanal and then centrifuged at 12,000 rpm for 10 min. The supernatant was analyzed by HPLC.
Supporting Information
| Supporting Information File 1: Spectroscopic data for compounds 1–3, HRMS–ESI data for compound 8, annotation of genes in the dat biosynthetic gene cluster, list of biological material, vectors, and primers used in this study. | ||
| Format: PDF | Size: 1.5 MB | Download |
Funding
This research was financially supported by the National Natural Science Foundation of China (82073738, 31972852, and U1702285), the Yunnan Provincial Science and Technology Department (2019FJ007 and 2019FA034), Biological Resources Program (KFJ-BRP-009), and Research Program of Frontier Sciences (QYZDB-SSW-SMC051), Yunnan Thousand Talents Plan Young Talent and Youth Innovation Promotion Association (2018424), Chinese Academy of Sciences.
References
-
Calì, V.; Spatafora, C.; Tringali, C. Stud. Nat. Prod. Chem. 2003, 29, 263–307. doi:10.1016/s1572-5995(03)80009-1
Return to citation in text: [1] -
Liu, J.-K. Chem. Rev. 2006, 106, 2209–2223. doi:10.1021/cr050248c
Return to citation in text: [1] -
Li, W.; Li, X.-B.; Lou, H.-X. J. Asian Nat. Prod. Res. 2018, 20, 1–13. doi:10.1080/10286020.2017.1381089
Return to citation in text: [1] -
Zhou, G.; Zhu, T.; Che, Q.; Zhang, G.; Li, D. Mar. Life Sci. Technol. 2022, 4, 62–73. doi:10.1007/s42995-021-00117-8
Return to citation in text: [1] -
Affleck, K.; Sidebottom, P. J.; Taylor, N. L.; Drake, C. S.; Todd, M.; Jowett, A.; Webb, G. J. Antibiot. 1999, 52, 89–95. doi:10.7164/antibiotics.52.89
Return to citation in text: [1] -
Wang, D.; Wang, Y.; Ouyang, Y.; Fu, P.; Zhu, W. J. Nat. Prod. 2019, 82, 3504–3508. doi:10.1021/acs.jnatprod.9b00963
Return to citation in text: [1] -
Belofsky, G. N.; Gloer, K. B.; Gloer, J. B.; Wicklow, D. T.; Dowd, P. F. J. Nat. Prod. 1998, 61, 1115–1119. doi:10.1021/np980188o
Return to citation in text: [1] -
Tian, S.-Z.; Pu, X.; Luo, G.; Zhao, L.-X.; Xu, L.-H.; Li, W.-J.; Luo, Y. J. Agric. Food Chem. 2013, 61, 3006–3012. doi:10.1021/jf400718w
Return to citation in text: [1] -
Quang, D. N.; Hashimoto, T.; Nukada, M.; Yamamoto, I.; Tanaka, M.; Asakawa, Y. Planta Med. 2003, 69, 1063–1066. doi:10.1055/s-2003-45159
Return to citation in text: [1] -
Liu, J.-K.; Hu, L.; Dong, Z.-J.; Hu, Q. Chem. Biodiversity 2004, 1, 601–605. doi:10.1002/cbdv.200490050
Return to citation in text: [1] -
Kuhnert, E.; Surup, F.; Herrmann, J.; Huch, V.; Müller, R.; Stadler, M. Phytochemistry 2015, 118, 68–73. doi:10.1016/j.phytochem.2015.08.004
Return to citation in text: [1] -
Lee, I.-K.; Jung, J.-Y.; Kim, Y.-S.; Rhee, M. H.; Yun, B.-S. Bioorg. Med. Chem. 2009, 17, 4674–4680. doi:10.1016/j.bmc.2009.04.064
Return to citation in text: [1] -
Kamigauchi, T.; Sakazaki, R.; Nagashima, K.; Kawamura, Y.; Yasuda, Y.; Matsushima, K.; Tani, H.; Takahashi, Y.; Ishii, K.; Suzuki, R.; Koizumi, K.; Nakai, H.; Ikenishi, Y.; Terui, Y. J. Antibiot. 1998, 51, 445–450. doi:10.7164/antibiotics.51.445
Return to citation in text: [1] -
Khanna, J. M.; Malone, M. H.; Euler, K. L.; Brady, L. R. J. Pharm. Sci. 1965, 54, 1016–1020. doi:10.1002/jps.2600540714
Return to citation in text: [1] -
Read, G.; Vining, L. C.; Haskins, R. H. Can. J. Chem. 1962, 40, 2357–2361. doi:10.1139/v62-359
Return to citation in text: [1] -
von Massow, F. Phytochemistry 1977, 16, 1695–1698. doi:10.1016/0031-9422(71)85072-0
Return to citation in text: [1] -
Braesel, J.; Götze, S.; Shah, F.; Heine, D.; Tauber, J.; Hertweck, C.; Tunlid, A.; Stallforth, P.; Hoffmeister, D. Chem. Biol. 2015, 22, 1325–1334. doi:10.1016/j.chembiol.2015.08.016
Return to citation in text: [1] [2] -
Fukuda, T.; Nagai, K.; Tomoda, H. J. Nat. Prod. 2012, 75, 2228–2231. doi:10.1021/np300428r
Return to citation in text: [1] -
Morisaki, Y.; Tsuji, Y.; Chujo, Y. Tetrahedron Lett. 2014, 55, 1631–1634. doi:10.1016/j.tetlet.2014.01.093
Return to citation in text: [1] -
Nakagawa, F.; Takahashi, S.; Naito, A.; Sato, S.; Iwabuchi, S.; Tamura, C. J. Antibiot. 1984, 37, 10–12. doi:10.7164/antibiotics.37.10
Return to citation in text: [1] -
Biggins, J. B.; Liu, X.; Feng, Z.; Brady, S. F. J. Am. Chem. Soc. 2011, 133, 1638–1641. doi:10.1021/ja1087369
Return to citation in text: [1] -
Lee, I.-K.; Yun, B.-S.; Cho, S.-M.; Kim, W.-G.; Kim, J.-P.; Ryoo, I.-J.; Koshino, H.; Yoo, I.-D. J. Nat. Prod. 1996, 59, 1090–1092. doi:10.1021/np960253z
Return to citation in text: [1] -
Feling, R.; Polborn, K.; Steglich, W.; Mühlbacher, J.; Bringmann, G. Tetrahedron 2001, 57, 7857–7863. doi:10.1016/s0040-4020(01)00761-x
Return to citation in text: [1] -
Eastwood, D. C.; Floudas, D.; Binder, M.; Majcherczyk, A.; Schneider, P.; Aerts, A.; Asiegbu, F. O.; Baker, S. E.; Barry, K.; Bendiksby, M.; Blumentritt, M.; Coutinho, P. M.; Cullen, D.; de Vries, R. P.; Gathman, A.; Goodell, B.; Henrissat, B.; Ihrmark, K.; Kauserud, H.; Kohler, A.; LaButti, K.; Lapidus, A.; Lavin, J. L.; Lee, Y.-H.; Lindquist, E.; Lilly, W.; Lucas, S.; Morin, E.; Murat, C.; Oguiza, J. A.; Park, J.; Pisabarro, A. G.; Riley, R.; Rosling, A.; Salamov, A.; Schmidt, O.; Schmutz, J.; Skrede, I.; Stenlid, J.; Wiebenga, A.; Xie, X.; Kües, U.; Hibbett, D. S.; Hoffmeister, D.; Högberg, N.; Martin, F.; Grigoriev, I. V.; Watkinson, S. C. Science 2011, 333, 762–765. doi:10.1126/science.1205411
Return to citation in text: [1] -
Shah, F.; Schwenk, D.; Nicolás, C.; Persson, P.; Hoffmeister, D.; Tunlid, A. Appl. Environ. Microbiol. 2015, 81, 8427–8433. doi:10.1128/aem.02312-15
Return to citation in text: [1] -
Zhu, J.; Chen, W.; Li, Y.-Y.; Deng, J.-J.; Zhu, D.-Y.; Duan, J.; Liu, Y.; Shi, G.-Y.; Xie, C.; Wang, H.-X.; Shen, Y.-M. Gene 2014, 546, 352–358. doi:10.1016/j.gene.2014.05.053
Return to citation in text: [1] [2] -
Sawitzke, J. A.; Thomason, L. C.; Bubunenko, M.; Li, X.; Costantino, N.; Court, D. L. Methods Enzymol. 2013, 533, 79–102. doi:10.1016/b978-0-12-420067-8.00007-6
Return to citation in text: [1]
| 27. | Sawitzke, J. A.; Thomason, L. C.; Bubunenko, M.; Li, X.; Costantino, N.; Court, D. L. Methods Enzymol. 2013, 533, 79–102. doi:10.1016/b978-0-12-420067-8.00007-6 |
| 26. | Zhu, J.; Chen, W.; Li, Y.-Y.; Deng, J.-J.; Zhu, D.-Y.; Duan, J.; Liu, Y.; Shi, G.-Y.; Xie, C.; Wang, H.-X.; Shen, Y.-M. Gene 2014, 546, 352–358. doi:10.1016/j.gene.2014.05.053 |
| 26. | Zhu, J.; Chen, W.; Li, Y.-Y.; Deng, J.-J.; Zhu, D.-Y.; Duan, J.; Liu, Y.; Shi, G.-Y.; Xie, C.; Wang, H.-X.; Shen, Y.-M. Gene 2014, 546, 352–358. doi:10.1016/j.gene.2014.05.053 |
| 1. | Calì, V.; Spatafora, C.; Tringali, C. Stud. Nat. Prod. Chem. 2003, 29, 263–307. doi:10.1016/s1572-5995(03)80009-1 |
| 2. | Liu, J.-K. Chem. Rev. 2006, 106, 2209–2223. doi:10.1021/cr050248c |
| 3. | Li, W.; Li, X.-B.; Lou, H.-X. J. Asian Nat. Prod. Res. 2018, 20, 1–13. doi:10.1080/10286020.2017.1381089 |
| 4. | Zhou, G.; Zhu, T.; Che, Q.; Zhang, G.; Li, D. Mar. Life Sci. Technol. 2022, 4, 62–73. doi:10.1007/s42995-021-00117-8 |
| 13. | Kamigauchi, T.; Sakazaki, R.; Nagashima, K.; Kawamura, Y.; Yasuda, Y.; Matsushima, K.; Tani, H.; Takahashi, Y.; Ishii, K.; Suzuki, R.; Koizumi, K.; Nakai, H.; Ikenishi, Y.; Terui, Y. J. Antibiot. 1998, 51, 445–450. doi:10.7164/antibiotics.51.445 |
| 24. | Eastwood, D. C.; Floudas, D.; Binder, M.; Majcherczyk, A.; Schneider, P.; Aerts, A.; Asiegbu, F. O.; Baker, S. E.; Barry, K.; Bendiksby, M.; Blumentritt, M.; Coutinho, P. M.; Cullen, D.; de Vries, R. P.; Gathman, A.; Goodell, B.; Henrissat, B.; Ihrmark, K.; Kauserud, H.; Kohler, A.; LaButti, K.; Lapidus, A.; Lavin, J. L.; Lee, Y.-H.; Lindquist, E.; Lilly, W.; Lucas, S.; Morin, E.; Murat, C.; Oguiza, J. A.; Park, J.; Pisabarro, A. G.; Riley, R.; Rosling, A.; Salamov, A.; Schmidt, O.; Schmutz, J.; Skrede, I.; Stenlid, J.; Wiebenga, A.; Xie, X.; Kües, U.; Hibbett, D. S.; Hoffmeister, D.; Högberg, N.; Martin, F.; Grigoriev, I. V.; Watkinson, S. C. Science 2011, 333, 762–765. doi:10.1126/science.1205411 |
| 25. | Shah, F.; Schwenk, D.; Nicolás, C.; Persson, P.; Hoffmeister, D.; Tunlid, A. Appl. Environ. Microbiol. 2015, 81, 8427–8433. doi:10.1128/aem.02312-15 |
| 9. | Quang, D. N.; Hashimoto, T.; Nukada, M.; Yamamoto, I.; Tanaka, M.; Asakawa, Y. Planta Med. 2003, 69, 1063–1066. doi:10.1055/s-2003-45159 |
| 10. | Liu, J.-K.; Hu, L.; Dong, Z.-J.; Hu, Q. Chem. Biodiversity 2004, 1, 601–605. doi:10.1002/cbdv.200490050 |
| 11. | Kuhnert, E.; Surup, F.; Herrmann, J.; Huch, V.; Müller, R.; Stadler, M. Phytochemistry 2015, 118, 68–73. doi:10.1016/j.phytochem.2015.08.004 |
| 12. | Lee, I.-K.; Jung, J.-Y.; Kim, Y.-S.; Rhee, M. H.; Yun, B.-S. Bioorg. Med. Chem. 2009, 17, 4674–4680. doi:10.1016/j.bmc.2009.04.064 |
| 17. | Braesel, J.; Götze, S.; Shah, F.; Heine, D.; Tauber, J.; Hertweck, C.; Tunlid, A.; Stallforth, P.; Hoffmeister, D. Chem. Biol. 2015, 22, 1325–1334. doi:10.1016/j.chembiol.2015.08.016 |
| 7. | Belofsky, G. N.; Gloer, K. B.; Gloer, J. B.; Wicklow, D. T.; Dowd, P. F. J. Nat. Prod. 1998, 61, 1115–1119. doi:10.1021/np980188o |
| 8. | Tian, S.-Z.; Pu, X.; Luo, G.; Zhao, L.-X.; Xu, L.-H.; Li, W.-J.; Luo, Y. J. Agric. Food Chem. 2013, 61, 3006–3012. doi:10.1021/jf400718w |
| 22. | Lee, I.-K.; Yun, B.-S.; Cho, S.-M.; Kim, W.-G.; Kim, J.-P.; Ryoo, I.-J.; Koshino, H.; Yoo, I.-D. J. Nat. Prod. 1996, 59, 1090–1092. doi:10.1021/np960253z |
| 5. | Affleck, K.; Sidebottom, P. J.; Taylor, N. L.; Drake, C. S.; Todd, M.; Jowett, A.; Webb, G. J. Antibiot. 1999, 52, 89–95. doi:10.7164/antibiotics.52.89 |
| 6. | Wang, D.; Wang, Y.; Ouyang, Y.; Fu, P.; Zhu, W. J. Nat. Prod. 2019, 82, 3504–3508. doi:10.1021/acs.jnatprod.9b00963 |
| 23. | Feling, R.; Polborn, K.; Steglich, W.; Mühlbacher, J.; Bringmann, G. Tetrahedron 2001, 57, 7857–7863. doi:10.1016/s0040-4020(01)00761-x |
| 18. | Fukuda, T.; Nagai, K.; Tomoda, H. J. Nat. Prod. 2012, 75, 2228–2231. doi:10.1021/np300428r |
| 20. | Nakagawa, F.; Takahashi, S.; Naito, A.; Sato, S.; Iwabuchi, S.; Tamura, C. J. Antibiot. 1984, 37, 10–12. doi:10.7164/antibiotics.37.10 |
| 17. | Braesel, J.; Götze, S.; Shah, F.; Heine, D.; Tauber, J.; Hertweck, C.; Tunlid, A.; Stallforth, P.; Hoffmeister, D. Chem. Biol. 2015, 22, 1325–1334. doi:10.1016/j.chembiol.2015.08.016 |
| 21. | Biggins, J. B.; Liu, X.; Feng, Z.; Brady, S. F. J. Am. Chem. Soc. 2011, 133, 1638–1641. doi:10.1021/ja1087369 |
| 15. | Read, G.; Vining, L. C.; Haskins, R. H. Can. J. Chem. 1962, 40, 2357–2361. doi:10.1139/v62-359 |
| 16. | von Massow, F. Phytochemistry 1977, 16, 1695–1698. doi:10.1016/0031-9422(71)85072-0 |
| 14. | Khanna, J. M.; Malone, M. H.; Euler, K. L.; Brady, L. R. J. Pharm. Sci. 1965, 54, 1016–1020. doi:10.1002/jps.2600540714 |
| 19. | Morisaki, Y.; Tsuji, Y.; Chujo, Y. Tetrahedron Lett. 2014, 55, 1631–1634. doi:10.1016/j.tetlet.2014.01.093 |
© 2022 Chen et al.; licensee Beilstein-Institut.
This is an open access article licensed under the terms of the Beilstein-Institut Open Access License Agreement (https://www.beilstein-journals.org/bjoc/terms), which is identical to the Creative Commons Attribution 4.0 International License (https://creativecommons.org/licenses/by/4.0). The reuse of material under this license requires that the author(s), source and license are credited. Third-party material in this article could be subject to other licenses (typically indicated in the credit line), and in this case, users are required to obtain permission from the license holder to reuse the material.