Abstract
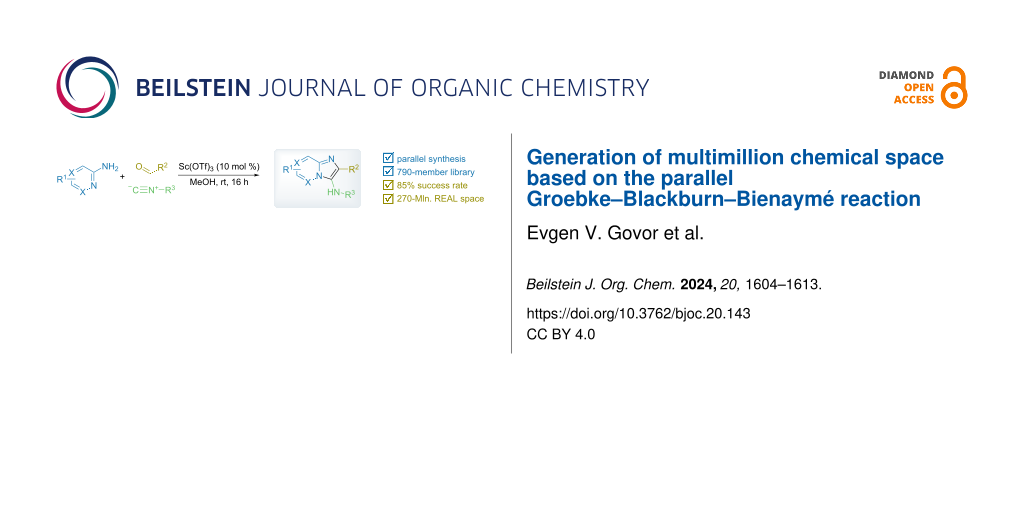
Parallel Groebke–Blackburn–Bienaymé reaction was evaluated as a source of multimillion chemically accessible chemical space. Two most popular classical protocols involving the use of Sc(OTf)3 and TsOH as the catalysts were tested on a broad substrate scope, and prevalence of the first method was clearly demonstrated. Furthermore, the scope and limitations of the procedure were established. A model 790-member library was obtained with 85% synthesis success rate. These results were used to generate a 271-Mln. readily accessible (REAL) heterocyclic chemical space mostly containing unique chemotypes, which was confirmed by comparative analysis with commercially available compound collections. Meanwhile, this chemical space contained 432 compounds that already showed biological activity according to the ChEMBL database.
Graphical Abstract
Introduction
Multicomponent reactions are widely recognized as a powerful source of biologically active compounds, in particular, for drug discovery purposes [1-6]. Isonitrile-based multicomponent reactions, such as the Groebke–Blackburn–Bienaymé (GBB) reaction, is an important tool in chemical synthesis providing easy access to a huge compound diversity and complexity [7-17]. Essentially, the GBB reaction is a three-component condensation of an α-amino heterocycle (e.g., 2-aminopyridine) 1, an aldehyde 2, and an isonitrile 3 providing the corresponding fused imidazoles (e.g., imidazo[1,2-a]pyridines) of general formula 4 (Scheme 1) [18-21]. Imidazo[1,2-a]pyridines and related heterocycles can be considered as privileged chemotypes in drug discovery: their representatives include the sedative drug zolpidem, disease-modifying antirheumatic agent upadacitinib, anticancer drug capmatinib, or risdiplam, a medication for spinal muscular atrophy (SMA) treatment (Figure 1) [22,23].
Scheme 1: Groebke–Blackburn–Bienaymé (GBB) reaction.
Scheme 1: Groebke–Blackburn–Bienaymé (GBB) reaction.
Figure 1: Marketed drugs comprising imidazo[1,2-a]azine scaffolds.
Figure 1: Marketed drugs comprising imidazo[1,2-a]azine scaffolds.
Over the last two decades, the GBB reaction has been studied in more than 200 research papers [21] and a number of works (including the original publication by Blackburn and coauthors [20]) described its parallel synthesis version [24-29]. Recently, we have shown that other (pseudo-)three-component reactions are very effective for the generation of synthetically tractable ultra-large chemical space [30-32]. Such virtual but readily accessible (REAL) compound libraries demonstrated excellent performance in early drug discovery programs when combined with modern computational tools such as high-throughput docking or machine learning [33-37]. In this work, we aimed at the implementation of the GBB reaction for the generation of such ultra-large chemical space, including experimental evaluation of the synthesis success rate (SSR, i.e., percentage of experiments that allowed obtaining the target library member in pure form) on a large set of starting materials.
Through the article, the compound numbering system common for the works on combinatorial chemistry was used: the starting materials used for the library generation were marked as 1{i}, 2{j}, and 3{k}, whereas the corresponding library members were denoted as 4{i,j,k}.
Results and Discussion
Library synthesis
Preliminary experiments on the parallel GBB reaction were performed with heterocyclic amines 1{1–430}, aldehydes 2{1–583}, and isonitriles 3{1–73} available from our stock (based on our previous in-house experience on isonitrile-based parallel reactions, electron-poor (hetero)aromatic isonitriles were not included in the study). According to Boltjes and Dömling, the following three catalysts were applied most often to promote the title reaction [21]: Sc(OTf)3 (described first in the original work by Blackburn and coauthors [20]), HClO4, and TsOH. We wanted to avoid the use of HClO4 in our parallel reaction set-up, so that only two remaining catalytic systems were evaluated. 580 library members were deliberately selected for both reaction conditions, and the corresponding experiments were performed (reactants at 1:1:1 ratio, 10 mol % of the catalyst, MeOH, rt, 16 h). It was found that TsOH, albeit being cheaper, demonstrated a poorer performance as the reaction promotor (62% SSR vs 67% for Sc(OTf)3; 34% average yield in both cases). This is especially apparent if the product yields are compared for 24 library members that we attempted to obtain by both methods (Figure 2).
![[1860-5397-20-143-2]](/bjoc/content/figures/1860-5397-20-143-2.png?scale=2.0&max-width=1024&background=FFFFFF)
Figure 2: Yields of library members 4 synthesized using both Sc(OTf)3 and TsOH as the catalysts.
Figure 2: Yields of library members 4 synthesized using both Sc(OTf)3 and TsOH as the catalysts.
These preliminary experiments also allowed establishing the following limitations of the method and excluding the corresponding reactants from further studies (Figure 3):
- α-aminoazoles of varied electronic nature (e.g., 3-aminopyrazole (1{1}), 3-aminoisoxazole (1{2}), 2-aminothiazole (1{3}), 2-amino-1,3,4-thiadiazole (1{4}), or 2-aminotetrazole (1{5}) demonstrated poor conversion to the target products;
- 2-aminopyrimidines either gave isomeric mixtures (e.g., parent compound 1{6}, alkyl-substituted derivatives 1{7} and 1{8}) or showed low conversion (halogenated derivatives 1{9–11});
- 4-aminopyrimidines 1{12–16} also demonstrated low conversion;
- pyridine derivatives with electron-withdrawing substituents (such as NO2 at C-3 or C-5 positions, as well as CN, SO2NH2, or C(O)NH2 at the C-3 atom) did not work (compounds 1{17–21});
- for pyridazine or pyrazine derivatives, even the presence of halogen atoms was sufficient to deactivate the substrate (e.g., compounds 1{22–24});
- a dialkylamino or alkoxy group at the C-6 position of pyridine derivatives also hampered the substrate’s reactivity (e.g., compounds 1{25–27}).
Figure 3: Amino heterocycles 1{1–27} demonstrating poor performance in the parallel GBB reaction.
Figure 3: Amino heterocycles 1{1–27} demonstrating poor performance in the parallel GBB reaction.
Some of these results (e.g., on the reactivity of aminopyrimidine derivatives) were in accordance with the previous literature data [21,29]. Meanwhile, electronic effects of the substituents in the amino heterocycle reported in the previous works were somewhat contradictory. Whereas for the NO2 group, lowering the reactivity has been documented, other electron-withdrawing groups were reported to be generally compatible with the GBB reaction [21]. Interference of dialkylamino or alkoxy groups at the C-6 position was also mentioned previously [29].
In addition to that, it was found that electron-poor aromatic aldehydes (e.g., 2{1–3}) did not work in the GBB reaction (Figure 4). This result is in accordance with the previous findings [21]. Notably, steric effects were not significant since o,o′-disubstituted aldehydes (e.g., 2{4–6}) displayed usual efficiency. As might be expected from our previous experience, 4-fluoro- and 4-chloro-2-fluoro-1-isocyanobenzenes (3{1} and 3{2}) showed poor performance and unsatisfactory results were also observed for isocyanocyclopropane (3{3}).
Figure 4: (Hetero)aromatic aldehydes 2{1–6} illustrating electronic and steric effects on the parallel GBB reaction.
Figure 4: (Hetero)aromatic aldehydes 2{1–6} illustrating electronic and steric effects on the parallel GBB re...
Using the guidelines described above, we have updated the reactant lists with additional representatives and excluded those demonstrating poor performance. Using the resulting sets of amino heterocycles 1{22–485}, aldehydes 2{4–867}, and isonitriles 3{4–78}, 892 library members 4{22–485,4–867,4–78} were deliberately selected and subjected to the parallel synthesis using the Sc(OTf)3-based protocol. As a result, 790 library members were obtained successfully (SSR = 85%, average yield of 37%) (Scheme 2).
Scheme 2: A) Parallel GBB reaction and B) examples of library members 4 obtained (relative configurations are shown).
Scheme 2: A) Parallel GBB reaction and B) examples of library members 4 obtained (relative configurations are...
Chemical space generation
Since validation of the GBB reaction showed that it is compatible with the main readily accessible chemical space criteria (at least 80% synthesizability) [30], we have aimed at the generation of the corresponding REAL space. For this purpose, 686 amino heterocycles 1, 3,927 aldehydes 2, and 107 isonitriles 3 complying with our general in-house reactivity/availability filters were subjected to virtual coupling using the limitations mentioned above. Additionally, combinations providing compounds with more than two chiral centers were not included. This resulted in 271,026,660 library members with nearly 85% expected synthetic accessibility according to the model experiments described in the previous section.
Distributions of the resulting chemical space over molecular weight (MW), 1-octanol–water partition coefficient logarithm (log P), H-bond acceptor/donor count (HAcc/HDon), fraction of sp3-hybrid carbon atoms (F(sp3)), and rotatable bond count (RotB) are shown in Figure 5. It is apparent that the GBB chemical space contains many drug-like (69,043,101 molecules, 25%) and “beyond-Ro5” compounds (75%) [38]. Furthermore, although the proposed approach cannot be considered lead-oriented, it may provide 12,122,351 lead-like compounds compliant with the “rule-of-four” (MW < 400, log P < 4) [39], and 1,383,298 with the even stricter Churcher’s rules (MW = 200–350, log P = −1 to 3) [40].
![[1860-5397-20-143-5]](/bjoc/content/figures/1860-5397-20-143-5.png?scale=2.0&max-width=1024&background=FFFFFF)
Figure 5: Physicochemical properties of the chemical space of 271 Mln. members obtained by virtual GBB reaction (MW – molecular weight; HAcc/HDon – H-bond acceptor/donor count; F(sp3) – fraction of sp3-hybrid carbon atoms; RotB – rotatable bond count); compounds complying with specific Lipinski/Veber rules (MW ≤ 500, log P ≤ 5, HDon ≤ 5, HAcc ≤ 10, RotB ≤ 10 [38,41]) as well as compounds with F(sp3) > 0.5 are highlighted in blue, the rest of the compounds are shown in yellow.
Figure 5: Physicochemical properties of the chemical space of 271 Mln. members obtained by virtual GBB reacti...
Next, we compared the GBB chemical space with common chemical databases (ChEMBL [42], PubChem [43], and ZINC15 [44]), as well as our stock screening compound collection [45]. Due to the enormous size of the databases, pairwise Tanimoto analysis was performed at the extended Bemis–Murcko scaffold level. First, extended Bemis–Murcko scaffolds were generated by cutting off the side chains of the molecules and retaining ring systems and linkers between them. After removal of duplicates, Tanimoto similarity coefficients were calculated for each pair of the molecules in the compared databases (T = 1 and 0 for similar and very dissimilar molecules, respectively). Average pairwise values for each molecule from the database of comparison are depicted in Figure 6. The mean values for all the databases were in the range of 0.44–0.45, which shows that despite there are some representatives that are similar to already known compounds, the generated GBB chemical space is unique as compared to the available compound collections.
![[1860-5397-20-143-6]](/bjoc/content/figures/1860-5397-20-143-6.png?scale=2.0&max-width=1024&background=FFFFFF)
Figure 6: Distribution of maximal values among pairwise-calculated Tanimoto similarities T (MFP2 fingerprints [46]) of extended Bemis–Murcko scaffolds for the generated chemical space members (5.60 Mln. scaffolds) to the extended Bemis–Murcko scaffolds of A) ChEMBL compounds (v. 33); B) PubChem compounds (due to the large size of the dataset, a preliminary clusterization was performed to achieve ca. 5-fold size reduction); C) ZINC15 drug-like compounds, and D) enamine’s stock screening collection. Average T values are shown by dotted lines.
Figure 6: Distribution of maximal values among pairwise-calculated Tanimoto similarities T (MFP2 fingerprints ...
This fact is even more apparent from the t-distributed stochastic neighbour embedding (t-SNE) analysis, a technique widely used for the dimension reduction in data visualization [47]. Due to the relatively high computational costs of this method, we randomly selected 50,000 compounds to represent each database. The dimension reduction algorithm uses molecular features as the starting inputs to generate a few coordinates (in this case, t-SNE1 and t-SNE2) reflecting the probability of the molecules to be similar. In this way, data visualization becomes possible since similar molecules will likely have close values of t-SNE1 and t-SNE2. As apparent from Figure 7, there is a small overlap between the GBB chemical space (yellow datapoints) with all four databases of comparison (blue data points), which is another confirmation of the GBB space uniqueness.
![[1860-5397-20-143-7]](/bjoc/content/figures/1860-5397-20-143-7.png?scale=2.0&max-width=1024&background=FFFFFF)
Figure 7: t-Distributed stochastic neighbor embedding (t-SNE) comparative analysis of 50,000 randomly selected molecules picked from the generated chemical space and A) ChEMBL compounds; B) PubChem compounds; C) ZINC15 compounds; and D) enamine’s stock screening collection.
Figure 7: t-Distributed stochastic neighbor embedding (t-SNE) comparative analysis of 50,000 randomly selecte...
Notably, 432 members of the generated GBB chemical space were already present in the ChEMBL database [42]. Among them, potent nonacidic farnesoid X receptor (FXR) modulators [48], 5-lipooxygenase (5-LO) inhibitors [49], soluble epoxide hydrolase (sEH) inhibitors [50], HIV-1 non-nucleoside reverse transcriptase inhibitors [51], or potential agents against visceral leishmaniasis [52] were found (Figure 8).
Figure 8: Some biologically active representatives of the generated GBB chemical space found in the ChEMBL database.
Figure 8: Some biologically active representatives of the generated GBB chemical space found in the ChEMBL da...
Conclusion
The Groebke–Blackburn–Bienaymé (GBB) reaction, a three-component condensation of amino heterocycles, aldehydes, and isonitriles, is a powerful tool for the combinatorial synthesis of compound libraries. We have shown that the Sc(OTf)3-catalyzed version of the reaction has a wide substrate applicability and established some limitations under the parallel synthesis conditions. In particular, while the method was applicable to a wide range of aminopyridines, pyrazines, and pyridazines, it worked poorly for aminoazoles, aminopyrimidines, substrates with strong electron-withdrawing groups, and substrates bearing additional dialkylamino or alkoxy substituents. The electronic nature was a major limiting factor for other two components of the reaction, the aldehyde and isonitrile, while the steric factor was found to be not significant. The protocol was used to prepare a 790-member compound library with 85% synthesis success rate. Furthermore, a readily available (REAL) chemical space comprising 271 Mln. members was generated. It was rich in both drug-like and “beyond rule-of-five” compounds and had considerable uniqueness as compared to the available collections (as was shown by Tanimoto similarity and t-distributed stochastic neighbor embedding (t-SNE) comparative analyses). Still, 432 members of the generated chemical space were found in the ChEMBL database, and some of them had high potency against various biological targets.
Supporting Information
The Supporting Information for this article contains structures of reactants 1–3, experimental details on the parallel synthesis of compound library 4, experimental section including compound characterization data, and copies of NMR spectra.
| Supporting Information File 1: Structures of reactants 1, 2, and 3. | ||
| Format: ZIP | Size: 6.9 MB | Download |
| Supporting Information File 2: Parallel synthesis of compound library 4. | ||
| Format: PDF | Size: 624.9 KB | Download |
| Supporting Information File 3: Experimental part. | ||
| Format: PDF | Size: 1.0 MB | Download |
| Supporting Information File 4: Copies of NMR spectra. | ||
| Format: PDF | Size: 3.0 MB | Download |
Acknowledgements
Dedicated to the memory of Kyrylo Burmagin and Yaroslav Mayboroda, students of Chemical Faculty at Taras Shevchenko National University of Kyiv, trainees at Enamine, and volunteer soldiers who died defending their country. The authors thank Mr. Oleksandr Liashuk and Dr. Anastasiia Hurieva for their help with manuscript preparation, Prof. Dr. Anrdiy A. Tolmachev for his encouragement and support, and all the brave people of Ukraine for making this publication possible.
Data Availability Statement
Most data that supports the findings of this study is available in the published article and/or the supporting information to this article. Additional data cannot be shared due to restrictions that apply to availability.
References
-
Dömling, A.; Wang, W.; Wang, K. Chem. Rev. 2012, 112, 3083–3135. doi:10.1021/cr100233r
Return to citation in text: [1] -
John, S. E.; Gulati, S.; Shankaraiah, N. Org. Chem. Front. 2021, 8, 4237–4287. doi:10.1039/d0qo01480j
Return to citation in text: [1] -
Graebin, C. S.; Ribeiro, F. V.; Rogério, K. R.; Kümmerle, A. E. Curr. Org. Synth. 2019, 16, 855–899. doi:10.2174/1570179416666190718153703
Return to citation in text: [1] -
Graziano, G.; Stefanachi, A.; Contino, M.; Prieto-Díaz, R.; Ligresti, A.; Kumar, P.; Scilimati, A.; Sotelo, E.; Leonetti, F. Int. J. Mol. Sci. 2023, 24, 6581. doi:10.3390/ijms24076581
Return to citation in text: [1] -
Younus, H. A.; Al-Rashida, M.; Hameed, A.; Uroos, M.; Salar, U.; Rana, S.; Khan, K. M. Expert Opin. Ther. Pat. 2021, 31, 267–289. doi:10.1080/13543776.2021.1858797
Return to citation in text: [1] -
Malinakova, H. C. Rep. Org. Chem. 2015, 5, 75–90. doi:10.2147/roc.s65115
Return to citation in text: [1] -
Dömling, A. Chem. Rev. 2006, 106, 17–89. doi:10.1021/cr0505728
Return to citation in text: [1] -
Rudick, J. G.; Shaabani, S.; Dömling, A. Front. Chem. (Lausanne, Switz.) 2020, 7, 918. doi:10.3389/fchem.2019.00918
Return to citation in text: [1] -
Akritopoulou-Zanze, I. Curr. Opin. Chem. Biol. 2008, 12, 324–331. doi:10.1016/j.cbpa.2008.02.004
Return to citation in text: [1] -
Russo, C.; Brunelli, F.; Cesare Tron, G.; Giustiniano, M. Chem. – Eur. J. 2023, 29, e202203150. doi:10.1002/chem.202203150
Return to citation in text: [1] -
Divyavani, C.; Padmaja, P.; Reddy, P. N. Curr. Org. Synth. 2024, 21, 140–165. doi:10.2174/1570179420666230330170845
Return to citation in text: [1] -
Mohlala, R. L.; Coyanis, E. M. Phys. Sci. Rev. 2024. doi:10.1515/psr-2022-0206
Return to citation in text: [1] -
Shaabani, A.; Hooshmand, S. E. RSC Adv. 2016, 6, 58142–58159. doi:10.1039/c6ra11701e
Return to citation in text: [1] -
Hulme, C.; Lee, Y.-S. Mol. Diversity 2008, 12, 1–15. doi:10.1007/s11030-008-9072-1
Return to citation in text: [1] -
Hulme, C.; Bienaymé, H.; Nixey, T.; Chenera, B.; Jones, W.; Tempest, P.; Smith, A. L. Methods Enzymol. 2003, 369, 469–496. doi:10.1016/s0076-6879(03)69024-5
Return to citation in text: [1] -
Fragkiadakis, M.; Anastasiou, P.-K.; Volyrakis, I.; Pantousas, A.; Stoumpos, C. C.; Neochoritis, C. G. Chem. Commun. 2023, 59, 14411–14414. doi:10.1039/d3cc04597h
Return to citation in text: [1] -
Fragkiadakis, M.; Anastasiou, P.-K.; Zingiridis, M.; Triantafyllou-Rundell, M. E.; Reyes Romero, A.; Stoumpos, C. C.; Neochoritis, C. G. J. Org. Chem. 2023, 88, 12709–12715. doi:10.1021/acs.joc.3c01379
Return to citation in text: [1] -
Groebke, K.; Weber, L.; Mehlin, F. Synlett 1998, 661–663. doi:10.1055/s-1998-1721
Return to citation in text: [1] -
Bienaymé, H.; Bouzid, K. Angew. Chem., Int. Ed. 1998, 37, 2234–2237. doi:10.1002/(sici)1521-3773(19980904)37:16<2234::aid-anie2234>3.3.co;2-i
Return to citation in text: [1] -
Blackburn, C.; Guan, B.; Fleming, P.; Shiosaki, K.; Tsai, S. Tetrahedron Lett. 1998, 39, 3635–3638. doi:10.1016/s0040-4039(98)00653-4
Return to citation in text: [1] [2] [3] -
Boltjes, A.; Dömling, A. Eur. J. Org. Chem. 2019, 7007–7049. doi:10.1002/ejoc.201901124
Return to citation in text: [1] [2] [3] [4] [5] [6] -
Wishart, D. S.; Feunang, Y. D.; Guo, A. C.; Lo, E. J.; Marcu, A.; Grant, J. R.; Sajed, T.; Johnson, D.; Li, C.; Sayeeda, Z.; Assempour, N.; Iynkkaran, I.; Liu, Y.; Maciejewski, A.; Gale, N.; Wilson, A.; Chin, L.; Cummings, R.; Le, D.; Pon, A.; Knox, C.; Wilson, M. Nucleic Acids Res. 2018, 46, D1074–D1082. doi:10.1093/nar/gkx1037
Return to citation in text: [1] -
Wishart, D. S.; Knox, C.; Guo, A. C.; Shrivastava, S.; Hassanali, M.; Stothard, P.; Chang, Z.; Woolsey, J. Nucleic Acids Res. 2006, 34, D668–D672. doi:10.1093/nar/gkj067
Return to citation in text: [1] -
An, Y.; Lee, J.; Seo, H.; Bae, S.; Kang, J.; Lee, J.; Kim, J.; Nam, M. H.; Song, M.; Hwang, G. T. Org. Lett. 2023, 25, 4445–4450. doi:10.1021/acs.orglett.3c01366
Return to citation in text: [1] -
Baenziger, M.; Durantie, E.; Mathes, C. Synthesis 2017, 49, 2266–2274. doi:10.1055/s-0036-1588130
Return to citation in text: [1] -
Shukla, N. M.; Salunke, D. B.; Yoo, E.; Mutz, C. A.; Balakrishna, R.; David, S. A. Bioorg. Med. Chem. 2012, 20, 5850–5863. doi:10.1016/j.bmc.2012.07.052
Return to citation in text: [1] -
Kolomiiets, O. V.; Tsygankov, A. V.; Kornet, M. N.; Brazhko, A. A.; Musatov, V. I.; Chebanov, V. A. Beilstein J. Org. Chem. 2023, 19, 727–735. doi:10.3762/bjoc.19.53
Return to citation in text: [1] -
Devi, N.; Rawal, R. K.; Singh, V. Tetrahedron 2015, 71, 183–232. doi:10.1016/j.tet.2014.10.032
Return to citation in text: [1] -
Gao, K.; Shaabani, S.; Xu, R.; Zarganes-Tzitzikas, T.; Gao, L.; Ahmadianmoghaddam, M.; Groves, M. R.; Dömling, A. RSC Med. Chem. 2021, 12, 809–818. doi:10.1039/d1md00087j
Return to citation in text: [1] [2] [3] -
Grygorenko, O. O.; Radchenko, D. S.; Dziuba, I.; Chuprina, A.; Gubina, K. E.; Moroz, Y. S. iScience 2020, 23, 101681. doi:10.1016/j.isci.2020.101681
Return to citation in text: [1] [2] -
Radchenko, D. S.; Naumchyk, V. S.; Dziuba, I.; Kyrylchuk, A. A.; Gubina, K. E.; Moroz, Y. S.; Grygorenko, O. O. Mol. Diversity 2022, 26, 993–1004. doi:10.1007/s11030-021-10218-2
Return to citation in text: [1] -
Savych, O.; Kuchkovska, Y. O.; Bogolyubsky, A. V.; Konovets, A. I.; Gubina, K. E.; Pipko, S. E.; Zhemera, A. V.; Grishchenko, A. V.; Khomenko, D. N.; Brovarets, V. S.; Doroschuk, R.; Moroz, Y. S.; Grygorenko, O. O. ACS Comb. Sci. 2019, 21, 635–642. doi:10.1021/acscombsci.9b00120
Return to citation in text: [1] -
Stein, R. M.; Kang, H. J.; McCorvy, J. D.; Glatfelter, G. C.; Jones, A. J.; Che, T.; Slocum, S.; Huang, X.-P.; Savych, O.; Moroz, Y. S.; Stauch, B.; Johansson, L. C.; Cherezov, V.; Kenakin, T.; Irwin, J. J.; Shoichet, B. K.; Roth, B. L.; Dubocovich, M. L. Nature 2020, 579, 609–614. doi:10.1038/s41586-020-2027-0
Return to citation in text: [1] -
Sadybekov, A. A.; Sadybekov, A. V.; Liu, Y.; Iliopoulos-Tsoutsouvas, C.; Huang, X.-P.; Pickett, J.; Houser, B.; Patel, N.; Tran, N. K.; Tong, F.; Zvonok, N.; Jain, M. K.; Savych, O.; Radchenko, D. S.; Nikas, S. P.; Petasis, N. A.; Moroz, Y. S.; Roth, B. L.; Makriyannis, A.; Katritch, V. Nature 2022, 601, 452–459. doi:10.1038/s41586-021-04220-9
Return to citation in text: [1] -
Gorgulla, C.; Boeszoermenyi, A.; Wang, Z.-F.; Fischer, P. D.; Coote, P. W.; Padmanabha Das, K. M.; Malets, Y. S.; Radchenko, D. S.; Moroz, Y. S.; Scott, D. A.; Fackeldey, K.; Hoffmann, M.; Iavniuk, I.; Wagner, G.; Arthanari, H. Nature 2020, 580, 663–668. doi:10.1038/s41586-020-2117-z
Return to citation in text: [1] -
Tropsha, A.; Isayev, O.; Varnek, A.; Schneider, G.; Cherkasov, A. Nat. Rev. Drug Discovery 2024, 23, 141–155. doi:10.1038/s41573-023-00832-0
Return to citation in text: [1] -
Gahbauer, S.; DeLeon, C.; Braz, J. M.; Craik, V.; Kang, H. J.; Wan, X.; Huang, X.-P.; Billesbølle, C. B.; Liu, Y.; Che, T.; Deshpande, I.; Jewell, M.; Fink, E. A.; Kondratov, I. S.; Moroz, Y. S.; Irwin, J. J.; Basbaum, A. I.; Roth, B. L.; Shoichet, B. K. Nat. Commun. 2023, 14, 8067. doi:10.1038/s41467-023-43506-6
Return to citation in text: [1] -
Lipinski, C. A.; Lombardo, F.; Dominy, B. W.; Feeney, P. J. Adv. Drug Delivery Rev. 1997, 23, 3–25. doi:10.1016/s0169-409x(96)00423-1
Return to citation in text: [1] [2] -
Grygorenko, O. O.; Volochnyuk, D. M.; Ryabukhin, S. V.; Judd, D. B. Chem. – Eur. J. 2020, 26, 1196–1237. doi:10.1002/chem.201903232
Return to citation in text: [1] -
Nadin, A.; Hattotuwagama, C.; Churcher, I. Angew. Chem., Int. Ed. 2012, 51, 1114–1122. doi:10.1002/anie.201105840
Return to citation in text: [1] -
Veber, D. F.; Johnson, S. R.; Cheng, H.-Y.; Smith, B. R.; Ward, K. W.; Kopple, K. D. J. Med. Chem. 2002, 45, 2615–2623. doi:10.1021/jm020017n
Return to citation in text: [1] -
Mendez, D.; Gaulton, A.; Bento, A. P.; Chambers, J.; De Veij, M.; Félix, E.; Magariños, M. P.; Mosquera, J. F.; Mutowo, P.; Nowotka, M.; Gordillo-Marañón, M.; Hunter, F.; Junco, L.; Mugumbate, G.; Rodriguez-Lopez, M.; Atkinson, F.; Bosc, N.; Radoux, C. J.; Segura-Cabrera, A.; Hersey, A.; Leach, A. R. Nucleic Acids Res. 2019, 47, D930–D940. doi:10.1093/nar/gky1075
Return to citation in text: [1] [2] -
Kim, S.; Chen, J.; Cheng, T.; Gindulyte, A.; He, J.; He, S.; Li, Q.; Shoemaker, B. A.; Thiessen, P. A.; Yu, B.; Zaslavsky, L.; Zhang, J.; Bolton, E. E. Nucleic Acids Res. 2021, 49, D1388–D1395. doi:10.1093/nar/gkaa971
Return to citation in text: [1] -
Sterling, T.; Irwin, J. J. J. Chem. Inf. Model. 2015, 55, 2324–2337. doi:10.1021/acs.jcim.5b00559
Return to citation in text: [1] -
Grygorenko, O. O. Eur. J. Org. Chem. 2021, 6474–6477. doi:10.1002/ejoc.202101210
Return to citation in text: [1] -
van der Maaten, L.; Hinton, G. J. Mach. Learn. Res. 2008, 9, 2579–2605.
Return to citation in text: [1] -
Rogers, D.; Hahn, M. J. Chem. Inf. Model. 2010, 50, 742–754. doi:10.1021/ci100050t
Return to citation in text: [1] -
Flesch, D.; Cheung, S.-Y.; Schmidt, J.; Gabler, M.; Heitel, P.; Kramer, J.; Kaiser, A.; Hartmann, M.; Lindner, M.; Lüddens-Dämgen, K.; Heering, J.; Lamers, C.; Lüddens, H.; Wurglics, M.; Proschak, E.; Schubert-Zsilavecz, M.; Merk, D. J. Med. Chem. 2017, 60, 7199–7205. doi:10.1021/acs.jmedchem.7b00903
Return to citation in text: [1] -
Hieke, M.; Rödl, C. B.; Wisniewska, J. M.; la Buscató, E.; Stark, H.; Schubert-Zsilavecz, M.; Steinhilber, D.; Hofmann, B.; Proschak, E. Bioorg. Med. Chem. Lett. 2012, 22, 1969–1975. doi:10.1016/j.bmcl.2012.01.038
Return to citation in text: [1] -
Meirer, K.; Rödl, C. B.; Wisniewska, J. M.; George, S.; Häfner, A.-K.; Buscató, E.; Klingler, F.-M.; Hahn, S.; Berressem, D.; Wittmann, S. K.; Steinhilber, D.; Hofmann, B.; Proschak, E. J. Med. Chem. 2013, 56, 1777–1781. doi:10.1021/jm301617j
Return to citation in text: [1] -
Bode, M. L.; Gravestock, D.; Moleele, S. S.; van der Westhuyzen, C. W.; Pelly, S. C.; Steenkamp, P. A.; Hoppe, H. C.; Khan, T.; Nkabinde, L. A. Bioorg. Med. Chem. 2011, 19, 4227–4237. doi:10.1016/j.bmc.2011.05.062
Return to citation in text: [1] -
Akao, Y.; Canan, S.; Cao, Y.; Condroski, K.; Engkvist, O.; Itono, S.; Kaki, R.; Kimura, C.; Kogej, T.; Nagaoka, K.; Naito, A.; Nakai, H.; Pairaudeau, G.; Radu, C.; Roberts, I.; Shimada, M.; Shum, D.; Watanabe, N.-a.; Xie, H.; Yonezawa, S.; Yoshida, O.; Yoshida, R.; Mowbray, C.; Perry, B. RSC Med. Chem. 2021, 12, 384–393. doi:10.1039/d0md00353k
Return to citation in text: [1]
| 48. | Flesch, D.; Cheung, S.-Y.; Schmidt, J.; Gabler, M.; Heitel, P.; Kramer, J.; Kaiser, A.; Hartmann, M.; Lindner, M.; Lüddens-Dämgen, K.; Heering, J.; Lamers, C.; Lüddens, H.; Wurglics, M.; Proschak, E.; Schubert-Zsilavecz, M.; Merk, D. J. Med. Chem. 2017, 60, 7199–7205. doi:10.1021/acs.jmedchem.7b00903 |
| 49. | Hieke, M.; Rödl, C. B.; Wisniewska, J. M.; la Buscató, E.; Stark, H.; Schubert-Zsilavecz, M.; Steinhilber, D.; Hofmann, B.; Proschak, E. Bioorg. Med. Chem. Lett. 2012, 22, 1969–1975. doi:10.1016/j.bmcl.2012.01.038 |
| 50. | Meirer, K.; Rödl, C. B.; Wisniewska, J. M.; George, S.; Häfner, A.-K.; Buscató, E.; Klingler, F.-M.; Hahn, S.; Berressem, D.; Wittmann, S. K.; Steinhilber, D.; Hofmann, B.; Proschak, E. J. Med. Chem. 2013, 56, 1777–1781. doi:10.1021/jm301617j |
| 1. | Dömling, A.; Wang, W.; Wang, K. Chem. Rev. 2012, 112, 3083–3135. doi:10.1021/cr100233r |
| 2. | John, S. E.; Gulati, S.; Shankaraiah, N. Org. Chem. Front. 2021, 8, 4237–4287. doi:10.1039/d0qo01480j |
| 3. | Graebin, C. S.; Ribeiro, F. V.; Rogério, K. R.; Kümmerle, A. E. Curr. Org. Synth. 2019, 16, 855–899. doi:10.2174/1570179416666190718153703 |
| 4. | Graziano, G.; Stefanachi, A.; Contino, M.; Prieto-Díaz, R.; Ligresti, A.; Kumar, P.; Scilimati, A.; Sotelo, E.; Leonetti, F. Int. J. Mol. Sci. 2023, 24, 6581. doi:10.3390/ijms24076581 |
| 5. | Younus, H. A.; Al-Rashida, M.; Hameed, A.; Uroos, M.; Salar, U.; Rana, S.; Khan, K. M. Expert Opin. Ther. Pat. 2021, 31, 267–289. doi:10.1080/13543776.2021.1858797 |
| 6. | Malinakova, H. C. Rep. Org. Chem. 2015, 5, 75–90. doi:10.2147/roc.s65115 |
| 21. | Boltjes, A.; Dömling, A. Eur. J. Org. Chem. 2019, 7007–7049. doi:10.1002/ejoc.201901124 |
| 21. | Boltjes, A.; Dömling, A. Eur. J. Org. Chem. 2019, 7007–7049. doi:10.1002/ejoc.201901124 |
| 22. | Wishart, D. S.; Feunang, Y. D.; Guo, A. C.; Lo, E. J.; Marcu, A.; Grant, J. R.; Sajed, T.; Johnson, D.; Li, C.; Sayeeda, Z.; Assempour, N.; Iynkkaran, I.; Liu, Y.; Maciejewski, A.; Gale, N.; Wilson, A.; Chin, L.; Cummings, R.; Le, D.; Pon, A.; Knox, C.; Wilson, M. Nucleic Acids Res. 2018, 46, D1074–D1082. doi:10.1093/nar/gkx1037 |
| 23. | Wishart, D. S.; Knox, C.; Guo, A. C.; Shrivastava, S.; Hassanali, M.; Stothard, P.; Chang, Z.; Woolsey, J. Nucleic Acids Res. 2006, 34, D668–D672. doi:10.1093/nar/gkj067 |
| 30. | Grygorenko, O. O.; Radchenko, D. S.; Dziuba, I.; Chuprina, A.; Gubina, K. E.; Moroz, Y. S. iScience 2020, 23, 101681. doi:10.1016/j.isci.2020.101681 |
| 18. | Groebke, K.; Weber, L.; Mehlin, F. Synlett 1998, 661–663. doi:10.1055/s-1998-1721 |
| 19. | Bienaymé, H.; Bouzid, K. Angew. Chem., Int. Ed. 1998, 37, 2234–2237. doi:10.1002/(sici)1521-3773(19980904)37:16<2234::aid-anie2234>3.3.co;2-i |
| 20. | Blackburn, C.; Guan, B.; Fleming, P.; Shiosaki, K.; Tsai, S. Tetrahedron Lett. 1998, 39, 3635–3638. doi:10.1016/s0040-4039(98)00653-4 |
| 21. | Boltjes, A.; Dömling, A. Eur. J. Org. Chem. 2019, 7007–7049. doi:10.1002/ejoc.201901124 |
| 21. | Boltjes, A.; Dömling, A. Eur. J. Org. Chem. 2019, 7007–7049. doi:10.1002/ejoc.201901124 |
| 7. | Dömling, A. Chem. Rev. 2006, 106, 17–89. doi:10.1021/cr0505728 |
| 8. | Rudick, J. G.; Shaabani, S.; Dömling, A. Front. Chem. (Lausanne, Switz.) 2020, 7, 918. doi:10.3389/fchem.2019.00918 |
| 9. | Akritopoulou-Zanze, I. Curr. Opin. Chem. Biol. 2008, 12, 324–331. doi:10.1016/j.cbpa.2008.02.004 |
| 10. | Russo, C.; Brunelli, F.; Cesare Tron, G.; Giustiniano, M. Chem. – Eur. J. 2023, 29, e202203150. doi:10.1002/chem.202203150 |
| 11. | Divyavani, C.; Padmaja, P.; Reddy, P. N. Curr. Org. Synth. 2024, 21, 140–165. doi:10.2174/1570179420666230330170845 |
| 12. | Mohlala, R. L.; Coyanis, E. M. Phys. Sci. Rev. 2024. doi:10.1515/psr-2022-0206 |
| 13. | Shaabani, A.; Hooshmand, S. E. RSC Adv. 2016, 6, 58142–58159. doi:10.1039/c6ra11701e |
| 14. | Hulme, C.; Lee, Y.-S. Mol. Diversity 2008, 12, 1–15. doi:10.1007/s11030-008-9072-1 |
| 15. | Hulme, C.; Bienaymé, H.; Nixey, T.; Chenera, B.; Jones, W.; Tempest, P.; Smith, A. L. Methods Enzymol. 2003, 369, 469–496. doi:10.1016/s0076-6879(03)69024-5 |
| 16. | Fragkiadakis, M.; Anastasiou, P.-K.; Volyrakis, I.; Pantousas, A.; Stoumpos, C. C.; Neochoritis, C. G. Chem. Commun. 2023, 59, 14411–14414. doi:10.1039/d3cc04597h |
| 17. | Fragkiadakis, M.; Anastasiou, P.-K.; Zingiridis, M.; Triantafyllou-Rundell, M. E.; Reyes Romero, A.; Stoumpos, C. C.; Neochoritis, C. G. J. Org. Chem. 2023, 88, 12709–12715. doi:10.1021/acs.joc.3c01379 |
| 29. | Gao, K.; Shaabani, S.; Xu, R.; Zarganes-Tzitzikas, T.; Gao, L.; Ahmadianmoghaddam, M.; Groves, M. R.; Dömling, A. RSC Med. Chem. 2021, 12, 809–818. doi:10.1039/d1md00087j |
| 33. | Stein, R. M.; Kang, H. J.; McCorvy, J. D.; Glatfelter, G. C.; Jones, A. J.; Che, T.; Slocum, S.; Huang, X.-P.; Savych, O.; Moroz, Y. S.; Stauch, B.; Johansson, L. C.; Cherezov, V.; Kenakin, T.; Irwin, J. J.; Shoichet, B. K.; Roth, B. L.; Dubocovich, M. L. Nature 2020, 579, 609–614. doi:10.1038/s41586-020-2027-0 |
| 34. | Sadybekov, A. A.; Sadybekov, A. V.; Liu, Y.; Iliopoulos-Tsoutsouvas, C.; Huang, X.-P.; Pickett, J.; Houser, B.; Patel, N.; Tran, N. K.; Tong, F.; Zvonok, N.; Jain, M. K.; Savych, O.; Radchenko, D. S.; Nikas, S. P.; Petasis, N. A.; Moroz, Y. S.; Roth, B. L.; Makriyannis, A.; Katritch, V. Nature 2022, 601, 452–459. doi:10.1038/s41586-021-04220-9 |
| 35. | Gorgulla, C.; Boeszoermenyi, A.; Wang, Z.-F.; Fischer, P. D.; Coote, P. W.; Padmanabha Das, K. M.; Malets, Y. S.; Radchenko, D. S.; Moroz, Y. S.; Scott, D. A.; Fackeldey, K.; Hoffmann, M.; Iavniuk, I.; Wagner, G.; Arthanari, H. Nature 2020, 580, 663–668. doi:10.1038/s41586-020-2117-z |
| 36. | Tropsha, A.; Isayev, O.; Varnek, A.; Schneider, G.; Cherkasov, A. Nat. Rev. Drug Discovery 2024, 23, 141–155. doi:10.1038/s41573-023-00832-0 |
| 37. | Gahbauer, S.; DeLeon, C.; Braz, J. M.; Craik, V.; Kang, H. J.; Wan, X.; Huang, X.-P.; Billesbølle, C. B.; Liu, Y.; Che, T.; Deshpande, I.; Jewell, M.; Fink, E. A.; Kondratov, I. S.; Moroz, Y. S.; Irwin, J. J.; Basbaum, A. I.; Roth, B. L.; Shoichet, B. K. Nat. Commun. 2023, 14, 8067. doi:10.1038/s41467-023-43506-6 |
| 20. | Blackburn, C.; Guan, B.; Fleming, P.; Shiosaki, K.; Tsai, S. Tetrahedron Lett. 1998, 39, 3635–3638. doi:10.1016/s0040-4039(98)00653-4 |
| 30. | Grygorenko, O. O.; Radchenko, D. S.; Dziuba, I.; Chuprina, A.; Gubina, K. E.; Moroz, Y. S. iScience 2020, 23, 101681. doi:10.1016/j.isci.2020.101681 |
| 31. | Radchenko, D. S.; Naumchyk, V. S.; Dziuba, I.; Kyrylchuk, A. A.; Gubina, K. E.; Moroz, Y. S.; Grygorenko, O. O. Mol. Diversity 2022, 26, 993–1004. doi:10.1007/s11030-021-10218-2 |
| 32. | Savych, O.; Kuchkovska, Y. O.; Bogolyubsky, A. V.; Konovets, A. I.; Gubina, K. E.; Pipko, S. E.; Zhemera, A. V.; Grishchenko, A. V.; Khomenko, D. N.; Brovarets, V. S.; Doroschuk, R.; Moroz, Y. S.; Grygorenko, O. O. ACS Comb. Sci. 2019, 21, 635–642. doi:10.1021/acscombsci.9b00120 |
| 21. | Boltjes, A.; Dömling, A. Eur. J. Org. Chem. 2019, 7007–7049. doi:10.1002/ejoc.201901124 |
| 29. | Gao, K.; Shaabani, S.; Xu, R.; Zarganes-Tzitzikas, T.; Gao, L.; Ahmadianmoghaddam, M.; Groves, M. R.; Dömling, A. RSC Med. Chem. 2021, 12, 809–818. doi:10.1039/d1md00087j |
| 24. | An, Y.; Lee, J.; Seo, H.; Bae, S.; Kang, J.; Lee, J.; Kim, J.; Nam, M. H.; Song, M.; Hwang, G. T. Org. Lett. 2023, 25, 4445–4450. doi:10.1021/acs.orglett.3c01366 |
| 25. | Baenziger, M.; Durantie, E.; Mathes, C. Synthesis 2017, 49, 2266–2274. doi:10.1055/s-0036-1588130 |
| 26. | Shukla, N. M.; Salunke, D. B.; Yoo, E.; Mutz, C. A.; Balakrishna, R.; David, S. A. Bioorg. Med. Chem. 2012, 20, 5850–5863. doi:10.1016/j.bmc.2012.07.052 |
| 27. | Kolomiiets, O. V.; Tsygankov, A. V.; Kornet, M. N.; Brazhko, A. A.; Musatov, V. I.; Chebanov, V. A. Beilstein J. Org. Chem. 2023, 19, 727–735. doi:10.3762/bjoc.19.53 |
| 28. | Devi, N.; Rawal, R. K.; Singh, V. Tetrahedron 2015, 71, 183–232. doi:10.1016/j.tet.2014.10.032 |
| 29. | Gao, K.; Shaabani, S.; Xu, R.; Zarganes-Tzitzikas, T.; Gao, L.; Ahmadianmoghaddam, M.; Groves, M. R.; Dömling, A. RSC Med. Chem. 2021, 12, 809–818. doi:10.1039/d1md00087j |
| 51. | Bode, M. L.; Gravestock, D.; Moleele, S. S.; van der Westhuyzen, C. W.; Pelly, S. C.; Steenkamp, P. A.; Hoppe, H. C.; Khan, T.; Nkabinde, L. A. Bioorg. Med. Chem. 2011, 19, 4227–4237. doi:10.1016/j.bmc.2011.05.062 |
| 20. | Blackburn, C.; Guan, B.; Fleming, P.; Shiosaki, K.; Tsai, S. Tetrahedron Lett. 1998, 39, 3635–3638. doi:10.1016/s0040-4039(98)00653-4 |
| 21. | Boltjes, A.; Dömling, A. Eur. J. Org. Chem. 2019, 7007–7049. doi:10.1002/ejoc.201901124 |
| 52. | Akao, Y.; Canan, S.; Cao, Y.; Condroski, K.; Engkvist, O.; Itono, S.; Kaki, R.; Kimura, C.; Kogej, T.; Nagaoka, K.; Naito, A.; Nakai, H.; Pairaudeau, G.; Radu, C.; Roberts, I.; Shimada, M.; Shum, D.; Watanabe, N.-a.; Xie, H.; Yonezawa, S.; Yoshida, O.; Yoshida, R.; Mowbray, C.; Perry, B. RSC Med. Chem. 2021, 12, 384–393. doi:10.1039/d0md00353k |
| 40. | Nadin, A.; Hattotuwagama, C.; Churcher, I. Angew. Chem., Int. Ed. 2012, 51, 1114–1122. doi:10.1002/anie.201105840 |
| 38. | Lipinski, C. A.; Lombardo, F.; Dominy, B. W.; Feeney, P. J. Adv. Drug Delivery Rev. 1997, 23, 3–25. doi:10.1016/s0169-409x(96)00423-1 |
| 39. | Grygorenko, O. O.; Volochnyuk, D. M.; Ryabukhin, S. V.; Judd, D. B. Chem. – Eur. J. 2020, 26, 1196–1237. doi:10.1002/chem.201903232 |
| 47. | Rogers, D.; Hahn, M. J. Chem. Inf. Model. 2010, 50, 742–754. doi:10.1021/ci100050t |
| 42. | Mendez, D.; Gaulton, A.; Bento, A. P.; Chambers, J.; De Veij, M.; Félix, E.; Magariños, M. P.; Mosquera, J. F.; Mutowo, P.; Nowotka, M.; Gordillo-Marañón, M.; Hunter, F.; Junco, L.; Mugumbate, G.; Rodriguez-Lopez, M.; Atkinson, F.; Bosc, N.; Radoux, C. J.; Segura-Cabrera, A.; Hersey, A.; Leach, A. R. Nucleic Acids Res. 2019, 47, D930–D940. doi:10.1093/nar/gky1075 |
| 45. | Grygorenko, O. O. Eur. J. Org. Chem. 2021, 6474–6477. doi:10.1002/ejoc.202101210 |
| 43. | Kim, S.; Chen, J.; Cheng, T.; Gindulyte, A.; He, J.; He, S.; Li, Q.; Shoemaker, B. A.; Thiessen, P. A.; Yu, B.; Zaslavsky, L.; Zhang, J.; Bolton, E. E. Nucleic Acids Res. 2021, 49, D1388–D1395. doi:10.1093/nar/gkaa971 |
| 44. | Sterling, T.; Irwin, J. J. J. Chem. Inf. Model. 2015, 55, 2324–2337. doi:10.1021/acs.jcim.5b00559 |
| 38. | Lipinski, C. A.; Lombardo, F.; Dominy, B. W.; Feeney, P. J. Adv. Drug Delivery Rev. 1997, 23, 3–25. doi:10.1016/s0169-409x(96)00423-1 |
| 41. | Veber, D. F.; Johnson, S. R.; Cheng, H.-Y.; Smith, B. R.; Ward, K. W.; Kopple, K. D. J. Med. Chem. 2002, 45, 2615–2623. doi:10.1021/jm020017n |
| 42. | Mendez, D.; Gaulton, A.; Bento, A. P.; Chambers, J.; De Veij, M.; Félix, E.; Magariños, M. P.; Mosquera, J. F.; Mutowo, P.; Nowotka, M.; Gordillo-Marañón, M.; Hunter, F.; Junco, L.; Mugumbate, G.; Rodriguez-Lopez, M.; Atkinson, F.; Bosc, N.; Radoux, C. J.; Segura-Cabrera, A.; Hersey, A.; Leach, A. R. Nucleic Acids Res. 2019, 47, D930–D940. doi:10.1093/nar/gky1075 |
© 2024 Govor et al.; licensee Beilstein-Institut.
This is an open access article licensed under the terms of the Beilstein-Institut Open Access License Agreement (https://www.beilstein-journals.org/bjoc/terms), which is identical to the Creative Commons Attribution 4.0 International License (https://creativecommons.org/licenses/by/4.0). The reuse of material under this license requires that the author(s), source and license are credited. Third-party material in this article could be subject to other licenses (typically indicated in the credit line), and in this case, users are required to obtain permission from the license holder to reuse the material.