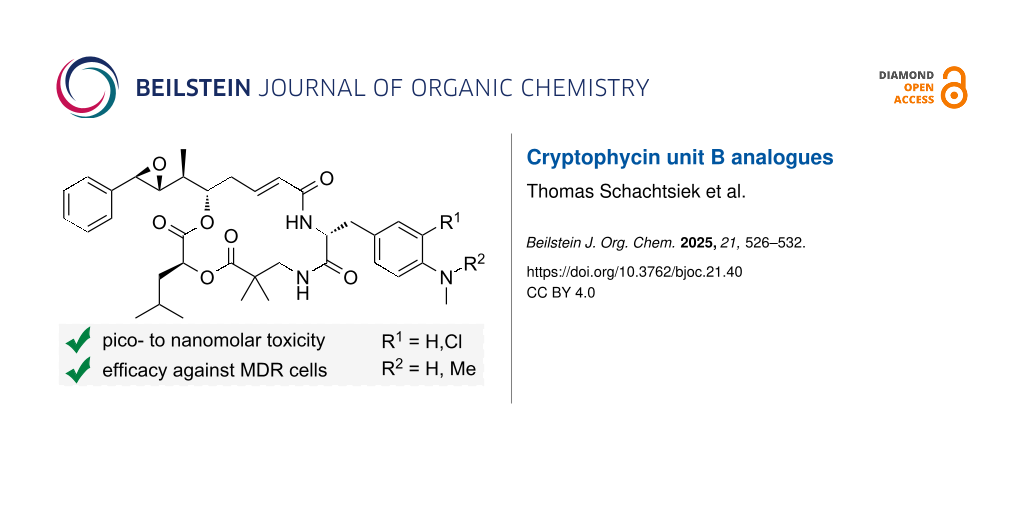
Abstract
Drug conjugates using toxic payloads are a promising approach for selectively combating cancer while sparing healthy tissue. The lack of highly cytotoxic and at the same time selective therapeutics against cancer is an ongoing challenge. Cryptophycins are a class of cyclic depsipeptides renowned for their high cytotoxicity in the picomolar range often combined with efficacy against multidrug-resistant tumour cell lines. However, cryptophycins failed as stand-alone drugs in cancer treatment, and their naturally occurring derivatives lack a covalent attachment handle. By making use of drug conjugates, toxic payloads such as cryptophycins can be selectively delivered to the target site. We present the synthesis of two conjugable cryptophycins with amino groups in unit B, representing potential payloads for drug conjugates particularly effective against multidrug-resistant cancers.
Graphical Abstract
Introduction
Cryptophycins emerged as highly potent cytotoxins for the use in targeted cancer therapy [1]. Originally discovered over three decades ago by isolation from cyanobacteria [2], their extraordinarily high cytotoxicity was apparent and attracted attention, not least because of their still high efficacy against multidrug-resistant (MDR) cells [3]. Cryptophycin-52, a synthetic development candidate by Eli Lilly based on the initially discovered cryptophycin-1, failed in clinical studies as a cytotoxic drug on its own due to severe side effects [4]. However, the embedment of cryptophycins as payloads in drug conjugates, increases specificity and minimises adverse effects [5,6]. Drug conjugates usually consist of three units, where the payload is connected to the homing device by a linker. For covalent attachment of the drug to the linker a suitable functional group is needed such as an amino, hydroxy, carboxy or sulfhydryl group [7]. Since the cryptophycins’ discovery, considerable efforts were made for the establishment of structure–activity relationship (SAR) studies between derivatisations of all four units (A to D) (Figure 1A) of cryptophycin-52 analogues with functional groups [8]. While many modifications of cryptophycin decrease the cytotoxicity drastically, some viable attachment points for conjugation were found, mainly including the para-phenyl position of unit A [9-11] or derivatisation of unit A’s epoxide moiety into a halohydrin (-glycinate) [5,12-17]. Cryptophycins modified at the α-position of unit C [18] and, most recently, various derivatives with modifications in unit D [19] were established as potent and conjugable payloads by our group. Modifications of unit B with conjugation handles are scarcely explored and mainly include the exchange of the para-methoxy group (Figure 1B). The sole exchange of the para-methoxy group of cryptophycin-52 (IC50 = 22 pM) by a hydroxy group reduces the cytotoxicity by approximately only one order of magnitude (IC50 = 0.52 nM) when tested on CCRF-CEM T lymphoblasts [20]. Loss of the meta-chloro substituent shows a similar trend. Functionalisation of the hydroxy group with ethylene glycol residues further decreases cytotoxicity, whereby this effect increases with increasing chain length [21]. The exchange by an amino group showed a similar trend, however, N,N-dimethylation of the amino group again increased cytotoxicity significantly [20]. Nevertheless, this cryptophycin might only be conjugated via an ammonium-based self-immolative linker [22]. We envisioned the synthesis of, firstly, m-chloro-p-(methylamino) derivative 1 to dissect the structure–activity relationship between the primary (IC50(CCRF-CEM) = 0.58 nM), secondary, and tertiary amine (IC50(CCRF-CEM) = 54 pM) derivatives [20]. Secondly, p-(dimethylamino) derivative 2 was synthesised to investigate the effect of N-alkylation on the non-chlorinated unit B derivatives.
Figure 1: A: Structure of cryptophycin-52. B: Cryptophycin-52 derivatives modified with conjugation handles in unit B. IC50 values given against (a) CCRF-CEM [20] and (b) KB-3-1 [21] cell lines. C: Cryptophycin-52 derivatives synthesised in this work.
Figure 1: A: Structure of cryptophycin-52. B: Cryptophycin-52 derivatives modified with conjugation handles i...
Results and Discussion
For the synthesis of unit B derivatives with amino groups instead of the naturally occurring methoxy group ᴅ-phenylalanine served as the fundamental substrate (Scheme 1). Nitration [23] followed by methyl ester formation and N-Boc-protection [24] provided nitroarene 5 in 40% yield over three steps. Reduction of the nitro group was performed with Pd/C and hydrogen to obtain aniline 6 in 98% yield, which served as a precursor for mono- and dimethylated unit B derivatives 7 and 8, respectively. While dimethylaniline 8 was obtained in good yield of 61% through reductive amination with excess formaldehyde and NaBH3CN as reductant, the selective installation of only one methyl group, providing monomethyl aniline 7, proved to be more troublesome. Either reductive amination using the same protocol, but under strict control of equivalents and pH, or Leuckart–Wallach-like reaction with ammonium formate and Pd/C [25] provided monomethylaniline 7 in 37% and 35% yield, respectively. The absolute structure of monomethylaniline 7 was confirmed by single-crystal X-ray diffraction measurements (Figure 2). Compound 7 crystallised in the monoclinic space group P21 with R1 = 0.0285 clearly showing the expected (R)-configuration with a Flack parameter of −0.07(6).
Scheme 1: Synthesis of modified unit B derivatives. a) HNO3, H2SO4, 0 °C, 5 h, 48% (isolated as monohydrate); b) SOCl2, MeOH, 0 °C, 90 min, then reflux, 17 h, 95%; c) Boc2O, NEt3, MeCN, H2O, rt, 22 h, 88%; d) H2, Pd/C, MeOH, rt, 25 h, 98%; e) either: formalin, NaBH3CN, HOAc, MeOH, rt, 20 min, 37%, or: formalin, (NH4)HCO2, Pd/C, MeOH, rt, 20 min, 35%; f) N-chlorosuccinimide, MeCN, reflux, 16 h, 77%; g) AllocCl, NaHCO3, CHCl3, H2O, rt, 5 h, 89%; h) LiOH·H2O, MeOH, tetrahydrofuran, H2O, 0 °C to rt, 3 h, 99%; i) formalin, NaBH3CN, MeCN, rt, 55 min, 61%; j) 1. TFA, CH2Cl2, 0 °C, 1.5 h; 2. acryloyl chloride, NEt3, CH2Cl2, 0 °C to rt, 19 h, 63% over two steps; k) LiOH·H2O, MeOH, tetrahydrofuran, H2O, 0 °C, 3.5 h, 82% with unconverted 12 (18 mol %).
Scheme 1: Synthesis of modified unit B derivatives. a) HNO3, H2SO4, 0 °C, 5 h, 48% (isolated as monohydrate);...
![[1860-5397-21-40-2]](/bjoc/content/figures/1860-5397-21-40-2.png?scale=2.0&max-width=1024&background=FFFFFF)
Figure 2: Molecular structure of Boc-ᴅ-Phe(4-NHMe)-OMe 7 as determined by single-crystal X-ray diffraction measurements. Thermal ellipsoids depicted at 50% probability.
Figure 2: Molecular structure of Boc-ᴅ-Phe(4-NHMe)-OMe 7 as determined by single-crystal X-ray diffraction me...
Starting from monomethylaniline derivative 7, the synthesis of the final building block was finalised by chlorination [20], Alloc protection and saponification [21] to obtain free acid 11. Exchange of the Boc protecting group of dimethylaniline 8 to an acryloyl substituent and subsequent saponification furnished dimethylamine building block 13.
For the macrocycle assembly, especially the ring closure, we decided on two different routes. While the cryptophycin containing a dimethylamino motif did not require an additional protecting group, ring closure was performed through alkene cross metathesis, which has been accomplished reliably and with good yields for other cryptophycins [11,26,27]. However, for the synthesis of a cryptophycin with a monomethylated amino group in unit B a suitable protecting group, i.e., allyloxycarbonyl (Alloc), must be used. Since the presence of this allylic double bond would most likely interfere with a clean reaction outcome after alkene cross metathesis, we decided for a more classical ring-closure strategy through macrolactamisation [21].
The syntheses of required unit A [28], C [29,30], and D [31] building blocks were accomplished as described previously. tert-Butyl-protected leucic acid 14 and Fmoc-β-aminopivalic acid (15) were connected by Steglich esterification (Scheme 2) and after cleavage of the tert-butyl ester group coupled to either one of the two unit A precursors 18 and 19 by Yamaguchi esterification. CDA fragments 20 and 21 were connected to unit B derivatives 11 and 13 through amidation which provided seco-cryptophycins 22 and 23 in yields of 49% and 38%, respectively. Starting from seco-cryptophycin 22 acidolysis was followed by macrolactamisation to obtain diol 24, albeit in poor yield of 11%. LC–MS analyses revealed that the low yield cannot be attributed to either incomplete acidolysis or incomplete conversion of the deprotected seco-cryptophycin during macrolactamisation. Rather, LC–MS analyses indicate the major presence of a fully deprotected and trifluoroacetylated seco-cryptophycin, most likely a TFA ester of one of the free hydroxy groups, a finding which might reason the comparably low yields (21–25%) of structurally similar unit B analogues reported earlier [21]. Contrary, diol 25 was obtained through Grubbs metathesis and subsequent acetonide cleavage in a superior yield of 76%.
Scheme 2: Synthesis of cryptophycin diols 24 and 25. a) EDC·HCl, DMAP, NEt3, CH2Cl2, 0 °C to rt, 22 h, 60%; b) TFA, CH2Cl2, 0 °C, 4 h, 100%; c) 2,4,6-trichlorobenzoyl chloride, DMAP, NEt3, THF, 0 °C or 0 °C to rt, 3h, tetrahydrofuran, 77% (for 20)/88% (for 21); d) 1. piperidine, N,N-dimethylformamide, rt, 1.5 h; 2. unit B (11 for 20 and 13 for 21), HOAt, NEt3, EDC·HCl, CH2Cl2, 0 °C to rt, 18 h, 49% (for 22)/38% (for 23); e) 1. TFA, CH2Cl2, 0 °C to rt, 2.5 h; 2. HATU, HOAt, N,N-dimethylformamide 0 °C for 2 h and at rt for 20 min, 11%; f) Grubbs II catalyst, CH2Cl2, reflux, 3 h, 84%; g) TFA, H2O, CH2Cl2, 0 °C to rt, 5 h, 91%. EDC·HCl = 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide hydrochloride; DMAP = 4-(dimethylamino)pyridine; TFA = trifluoroacetic acid; HOAt = 1-hydroxy-7-azabenzotriazole; HATU = 1-[bis(dimethylamino)methyliumyl]-1H-1,2,3-triazolo[4,5-b]pyridinium 3-oxide hexafluorophosphate.
Scheme 2: Synthesis of cryptophycin diols 24 and 25. a) EDC·HCl, DMAP, NEt3, CH2Cl2, 0 °C to rt, 22 h, 60%; b...
The finalising steps to obtain epoxides 26 and 2 (Scheme 3) were a diol–epoxide transformation [11,19,32], including firstly the formation of a cyclic orthoester, secondly the formation of a bromohydrin formate, and thirdly ring closure to obtain the epoxides 26 and 2 in 89% and 14% yield over three steps each, respectively.
Scheme 3: Three-step diol–epoxide transformation starting from diols 24 and 25. a) (MeO)3CH, pyridinium p-toluenesulfonate, CH2Cl2, rt, 3 h; b) AcBr, CH2Cl2, rt, 3–4 h; c) K2CO3, MeOCH2CH2OMe, HOCH2CH2OH, rt, 5 min, 89% (for 26)/14% (for 2), over three steps each; d) Pd(PPh3)4, morpholine, CH2Cl2, rt, 90 min, 74%.
Scheme 3: Three-step diol–epoxide transformation starting from diols 24 and 25. a) (MeO)3CH, pyridinium p-tol...
While the synthesis of tertiary amine 2 was finished, final Alloc cleavage from 26 under Tsuji–Trost-like conditions [19] provided secondary amine 1 in 74% yield.
The cytotoxicity of Alloc-protected cryptophycin 26 and both free amines 1 and 2 were tested against the human cervix carcinoma cell line KB-3-1 and its MDR subclone KB-V1 (Table 1) [33,34].
The m-chloro-p-(methylamino) derivative 1 showed high cytotoxicity with IC50 (KB-3-1) = 313 pM, which is perfectly between the values of the primary (IC50 (CCRF-CEM) = 580 pM) and tertiary (IC50 (CCRF-CEM) = 54 pM) amine derivatives (Figure 1B) [20,35], showing a strict increase in cytotoxicity with increasing degree of aniline methylation. The same trend was also observed for unit D derivatives modified with amino groups [19] and likely hints to favoured hydrophobic interactions around the binding pocket at unit B and D. In comparison, the cytotoxicity of Alloc-protected amine 26 decreased significantly (IC50 (KB-3-1) = 374 nM), which could be due to higher steric bulk as this is known to be less well tolerated; cf. unit B p-alkyloxy derivatives (Figure 1B) [21]. Tertiary amine 2 showed IC50 (KB-3-1) = 6.36 nM which is similar to its non-alkylated congener (IC50 (CCRF-CEM) = 10.11 nM) but about two orders of magnitude higher than its chlorinated derivative (IC50 (CCRF-CEM) = 54 pM) (Figure 1B) [20]. Notably, both free amines still show nanomolar activity against the MDR cell line KB-V1 (Table 1) with resistance factors FR (being the ratio between IC50 (KB-V1) and IC50 (KB-3-1)) of 25 (1) and 34 (2), thus being comparably much more potent against KB-V1 cells than for example the unit D analogues (FR ≈ 103) [19].
Conclusion
In summary, we synthesised two new conjugable cryptophycin analogues 1 and 2 with amino groups in unit B, by utilising either macrolactamisation or Grubbs metathesis for ring closure. These synthetic routes should generally allow access to diverse other unit B cryptophycin analogues. Both cryptophycins 1 and 2 showed high cytotoxicity with 313 pM (1) and 6.36 nM (2) and outstandingly low resistance factors. Furthermore, the new cryptophycin 1 confirms the correlation between degree of alkylation and cytotoxicity of m-chloro-p-amino unit B derivatives. Since MDR is responsible for over 90% of deaths in cancer patients and MDR-selective therapeutics are lacking [36], we expect that cryptophycins 1 and 2 will be considered as viable cytotoxins for the use in targeted tumour therapy, especially against MDR cancers.
Supporting Information
The supplementary crystallographic data for this article can be obtained free of charge from the Cambridge Crystallographic Data Centre (CCDC) via https://www.ccdc.cam.ac.uk/structures under reference number CCDC 2400694 for Boc-ᴅ-Phe(4-NHMe)-OMe (7).
| Supporting Information File 1: Experimental section, NMR spectra and cellular proliferation assays. | ||
| Format: PDF | Size: 4.6 MB | Download |
Data Availability Statement
All data that supports the findings of this study is available in the published article and/or the supporting information of this article.
References
-
Verma, V. A.; Pillow, T. H.; DePalatis, L.; Li, G.; Phillips, G. L.; Polson, A. G.; Raab, H. E.; Spencer, S.; Zheng, B. Bioorg. Med. Chem. Lett. 2015, 25, 864–868. doi:10.1016/j.bmcl.2014.12.070
Return to citation in text: [1] -
Schwartz, R. E.; Hirsch, C. F.; Sesin, D. F.; Flor, J. E.; Chartrain, M.; Fromtling, R. E.; Harris, G. H.; Salvatore, M. J.; Liesch, J. M.; Yudin, K. J. Ind. Microbiol. 1990, 5, 113–123. doi:10.1007/bf01573860
Return to citation in text: [1] -
Wagner, M. M.; Paul, D. C.; Shih, C.; Jordan, M. A.; Wilson, L.; Williams, D. C. Cancer Chemother. Pharmacol. 1999, 43, 115–125. doi:10.1007/s002800050871
Return to citation in text: [1] -
D'Agostino, G.; Del Campo, J.; Mellado, B.; Izquierdo, M. A.; Minarik, T.; Cirri, L.; Marini, L.; Perez-Gracia, J. L.; Scambia, G. Int. J. Gynecol. Cancer 2006, 16, 71–76. doi:10.1136/ijgc-00009577-200601000-00012
Return to citation in text: [1] -
Lai, Q.; Wu, M.; Wang, R.; Lai, W.; Tao, Y.; Lu, Y.; Wang, Y.; Yu, L.; Zhang, R.; Peng, Y.; Jiang, X.; Fu, Y.; Wang, X.; Zhang, Z.; Guo, C.; Liao, W.; Zhang, Y.; Kang, T.; Chen, H.; Yao, Y.; Gou, L.; Yang, J. Eur. J. Med. Chem. 2020, 199, 112364. doi:10.1016/j.ejmech.2020.112364
Return to citation in text: [1] [2] -
Steinkuehler, C. M.; Gallinari, P. M.; Osswald, B.; Sewald, N.; Ritzefeld, M.; Frese, M. Cryptophycin-based antibody-drug conjugates with novel self-immolative linkers. WO. Pat. Appl. WO2016146638A1, Sept 22, 2016.
Return to citation in text: [1] -
McCombs, J. R.; Owen, S. C. AAPS J. 2015, 17, 339–351. doi:10.1208/s12248-014-9710-8
Return to citation in text: [1] -
Weiss, C.; Figueras, E.; Borbely, A. N.; Sewald, N. J. Pept. Sci. 2017, 23, 514–531. doi:10.1002/psc.3015
Return to citation in text: [1] -
Bouchard, H.; Brun, M.-P.; Hubert, P. Novel peptidic linkers and cryptophycin conjugates, their preparation and their therapeutic use. U.S. Pat. Appl. US20210228726A1, July 29, 2021.
Return to citation in text: [1] -
McGonigle, S.; Majumder, U.; Custar, D. W.; Wu, J.; Postema, M. H. D. Peptide drug conjugates. WO Pat. Appl. WO2017136769A1, Aug 10, 2017.
Return to citation in text: [1] -
Eißler, S.; Bogner, T.; Nahrwold, M.; Sewald, N. Chem. – Eur. J. 2009, 15, 11273–11287. doi:10.1002/chem.200901750
Return to citation in text: [1] [2] [3] -
Figueras, E.; Martins, A.; Borbély, A.; Le Joncour, V.; Cordella, P.; Perego, R.; Modena, D.; Pagani, P.; Esposito, S.; Auciello, G.; Frese, M.; Gallinari, P.; Laakkonen, P.; Steinkühler, C.; Sewald, N. Pharmaceutics 2019, 11, 220. doi:10.3390/pharmaceutics11050220
Return to citation in text: [1] -
Anselmi, M.; Borbély, A.; Figueras, E.; Michalek, C.; Kemker, I.; Gentilucci, L.; Sewald, N. Chem. – Eur. J. 2021, 27, 1015–1022. doi:10.1002/chem.202003471
Return to citation in text: [1] -
Georg, G. I.; Ali, S. M.; Stella, V. J.; Waugh, W. N.; Himes, R. H. Bioorg. Med. Chem. Lett. 1998, 8, 1959–1962. doi:10.1016/s0960-894x(98)00356-4
Return to citation in text: [1] -
Cazzamalli, S.; Figueras, E.; Pethő, L.; Borbély, A.; Steinkühler, C.; Neri, D.; Sewald, N. ACS Omega 2018, 3, 14726–14731. doi:10.1021/acsomega.8b02350
Return to citation in text: [1] -
Borbély, A.; Figueras, E.; Martins, A.; Esposito, S.; Auciello, G.; Monteagudo, E.; Di Marco, A.; Summa, V.; Cordella, P.; Perego, R.; Kemker, I.; Frese, M.; Gallinari, P.; Steinkühler, C.; Sewald, N. Pharmaceutics 2019, 11, 151. doi:10.3390/pharmaceutics11040151
Return to citation in text: [1] -
Borbély, A.; Figueras, E.; Martins, A.; Bodero, L.; Raposo Moreira Dias, A.; López Rivas, P.; Pina, A.; Arosio, D.; Gallinari, P.; Frese, M.; Steinkühler, C.; Gennari, C.; Piarulli, U.; Sewald, N. ChemistryOpen 2019, 8, 737–742. doi:10.1002/open.201900110
Return to citation in text: [1] -
Nahrwold, M.; Weiß, C.; Bogner, T.; Mertink, F.; Conradi, J.; Sammet, B.; Palmisano, R.; Royo Gracia, S.; Preuße, T.; Sewald, N. J. Med. Chem. 2013, 56, 1853–1864. doi:10.1021/jm301346z
Return to citation in text: [1] -
Dessin, C.; Schachtsiek, T.; Voss, J.; Abel, A.-C.; Neumann, B.; Stammler, H.-G.; Prota, A. E.; Sewald, N. Angew. Chem., Int. Ed. 2024, 63, e202416210. doi:10.1002/anie.202416210
Return to citation in text: [1] [2] [3] [4] [5] -
Patel, V. F.; Andis, S. L.; Kennedy, J. H.; Ray, J. E.; Schultz, R. M. J. Med. Chem. 1999, 42, 2588–2603. doi:10.1021/jm980706s
Return to citation in text: [1] [2] [3] [4] [5] [6] [7] -
Figueras, E.; Borbély, A.; Ismail, M.; Frese, M.; Sewald, N. Beilstein J. Org. Chem. 2018, 14, 1281–1286. doi:10.3762/bjoc.14.109
Return to citation in text: [1] [2] [3] [4] [5] [6] -
Staben, L. R.; Koenig, S. G.; Lehar, S. M.; Vandlen, R.; Zhang, D.; Chuh, J.; Yu, S.-F.; Ng, C.; Guo, J.; Liu, Y.; Fourie-O'Donohue, A.; Go, M.; Linghu, X.; Segraves, N. L.; Wang, T.; Chen, J.; Wei, B.; Phillips, G. D. L.; Xu, K.; Kozak, K. R.; Mariathasan, S.; Flygare, J. A.; Pillow, T. H. Nat. Chem. 2016, 8, 1112–1119. doi:10.1038/nchem.2635
Return to citation in text: [1] -
Wang, L.; Hisano, W.; Murai, Y.; Sakurai, M.; Muto, Y.; Ikemoto, H.; Okamoto, M.; Murotani, T.; Isoda, R.; Kim, D.; Sakihama, Y.; Sitepu, I. R.; Hashidoko, Y.; Hatanaka, Y.; Hashimoto, M. Molecules 2013, 18, 8393–8401. doi:10.3390/molecules18078393
Return to citation in text: [1] -
Arns, S. P.; Jaquith, J. B.; Baudelet, D. J.; Simonson, E. R.; Liggins, R. T.; Kumar, N. S.; Hsieh, T. H. H. Somatostatin receptor antagonist compounds and methods of using the same. WO Patent WO2017/136943, Aug 17, 2017.
Return to citation in text: [1] -
Byun, E.; Hong, B.; De Castro, K. A.; Lim, M.; Rhee, H. J. Org. Chem. 2007, 72, 9815–9817. doi:10.1021/jo701503q
Return to citation in text: [1] -
Tripathy, N. K.; Georg, G. I. Tetrahedron Lett. 2004, 45, 5309–5311. doi:10.1016/j.tetlet.2004.04.164
Return to citation in text: [1] -
Bendelac, A.; Benedetti, F.; Doublet-Decabras, V.; Lokovi, R.; Decalogne, F.; Bigot, A. Org. Process Res. Dev. 2022, 26, 2145–2154. doi:10.1021/acs.oprd.2c00080
Return to citation in text: [1] -
Eissler, S.; Nahrwold, M.; Neumann, B.; Stammler, H.-G.; Sewald, N. Org. Lett. 2007, 9, 817–819. doi:10.1021/ol063032l
Return to citation in text: [1] -
Sammet, B.; Bogner, T.; Nahrwold, M.; Weiss, C.; Sewald, N. J. Org. Chem. 2010, 75, 6953–6960. doi:10.1021/jo101563s
Return to citation in text: [1] -
Nahrwold, M.; Bogner, T.; Eissler, S.; Verma, S.; Sewald, N. Org. Lett. 2010, 12, 1064–1067. doi:10.1021/ol1000473
Return to citation in text: [1] -
Weide, T.; Arve, L.; Prinz, H.; Waldmann, H.; Kessler, H. Bioorg. Med. Chem. Lett. 2006, 16, 59–63. doi:10.1016/j.bmcl.2005.09.051
Return to citation in text: [1] -
Kolb, H. C.; Sharpless, K. B. Tetrahedron 1992, 48, 10515–10530. doi:10.1016/s0040-4020(01)88349-6
Return to citation in text: [1] -
Akiyama, S.-i.; Fojo, A.; Hanover, J. A.; Pastan, I.; Gottesman, M. M. Somatic Cell Mol. Genet. 1985, 11, 117–126. doi:10.1007/bf01534700
Return to citation in text: [1] -
Shen, D. W.; Cardarelli, C.; Hwang, J.; Cornwell, M.; Richert, N.; Ishii, S.; Pastan, I.; Gottesman, M. M. J. Biol. Chem. 1986, 261, 7762–7770. doi:10.1016/s0021-9258(19)57466-x
Return to citation in text: [1] -
Although the tissue type of the KB-3-1 cell line (cervical carcinoma) and the CCRF-CEM cell line (T lymphoblasts) are different, there is generally only a very small difference in the IC50 values for many different cryptophycins (see Sewald et al., [8]), so that we assume good comparability of the IC50 values between the two cell lines.
Return to citation in text: [1] -
Bukowski, K.; Kciuk, M.; Kontek, R. Int. J. Mol. Sci. 2020, 21, 3233. doi:10.3390/ijms21093233
Return to citation in text: [1]
| 31. | Weide, T.; Arve, L.; Prinz, H.; Waldmann, H.; Kessler, H. Bioorg. Med. Chem. Lett. 2006, 16, 59–63. doi:10.1016/j.bmcl.2005.09.051 |
| 21. | Figueras, E.; Borbély, A.; Ismail, M.; Frese, M.; Sewald, N. Beilstein J. Org. Chem. 2018, 14, 1281–1286. doi:10.3762/bjoc.14.109 |
| 11. | Eißler, S.; Bogner, T.; Nahrwold, M.; Sewald, N. Chem. – Eur. J. 2009, 15, 11273–11287. doi:10.1002/chem.200901750 |
| 19. | Dessin, C.; Schachtsiek, T.; Voss, J.; Abel, A.-C.; Neumann, B.; Stammler, H.-G.; Prota, A. E.; Sewald, N. Angew. Chem., Int. Ed. 2024, 63, e202416210. doi:10.1002/anie.202416210 |
| 32. | Kolb, H. C.; Sharpless, K. B. Tetrahedron 1992, 48, 10515–10530. doi:10.1016/s0040-4020(01)88349-6 |
| 1. | Verma, V. A.; Pillow, T. H.; DePalatis, L.; Li, G.; Phillips, G. L.; Polson, A. G.; Raab, H. E.; Spencer, S.; Zheng, B. Bioorg. Med. Chem. Lett. 2015, 25, 864–868. doi:10.1016/j.bmcl.2014.12.070 |
| 5. | Lai, Q.; Wu, M.; Wang, R.; Lai, W.; Tao, Y.; Lu, Y.; Wang, Y.; Yu, L.; Zhang, R.; Peng, Y.; Jiang, X.; Fu, Y.; Wang, X.; Zhang, Z.; Guo, C.; Liao, W.; Zhang, Y.; Kang, T.; Chen, H.; Yao, Y.; Gou, L.; Yang, J. Eur. J. Med. Chem. 2020, 199, 112364. doi:10.1016/j.ejmech.2020.112364 |
| 6. | Steinkuehler, C. M.; Gallinari, P. M.; Osswald, B.; Sewald, N.; Ritzefeld, M.; Frese, M. Cryptophycin-based antibody-drug conjugates with novel self-immolative linkers. WO. Pat. Appl. WO2016146638A1, Sept 22, 2016. |
| 22. | Staben, L. R.; Koenig, S. G.; Lehar, S. M.; Vandlen, R.; Zhang, D.; Chuh, J.; Yu, S.-F.; Ng, C.; Guo, J.; Liu, Y.; Fourie-O'Donohue, A.; Go, M.; Linghu, X.; Segraves, N. L.; Wang, T.; Chen, J.; Wei, B.; Phillips, G. D. L.; Xu, K.; Kozak, K. R.; Mariathasan, S.; Flygare, J. A.; Pillow, T. H. Nat. Chem. 2016, 8, 1112–1119. doi:10.1038/nchem.2635 |
| 19. | Dessin, C.; Schachtsiek, T.; Voss, J.; Abel, A.-C.; Neumann, B.; Stammler, H.-G.; Prota, A. E.; Sewald, N. Angew. Chem., Int. Ed. 2024, 63, e202416210. doi:10.1002/anie.202416210 |
| 4. | D'Agostino, G.; Del Campo, J.; Mellado, B.; Izquierdo, M. A.; Minarik, T.; Cirri, L.; Marini, L.; Perez-Gracia, J. L.; Scambia, G. Int. J. Gynecol. Cancer 2006, 16, 71–76. doi:10.1136/ijgc-00009577-200601000-00012 |
| 20. | Patel, V. F.; Andis, S. L.; Kennedy, J. H.; Ray, J. E.; Schultz, R. M. J. Med. Chem. 1999, 42, 2588–2603. doi:10.1021/jm980706s |
| 36. | Bukowski, K.; Kciuk, M.; Kontek, R. Int. J. Mol. Sci. 2020, 21, 3233. doi:10.3390/ijms21093233 |
| 3. | Wagner, M. M.; Paul, D. C.; Shih, C.; Jordan, M. A.; Wilson, L.; Williams, D. C. Cancer Chemother. Pharmacol. 1999, 43, 115–125. doi:10.1007/s002800050871 |
| 21. | Figueras, E.; Borbély, A.; Ismail, M.; Frese, M.; Sewald, N. Beilstein J. Org. Chem. 2018, 14, 1281–1286. doi:10.3762/bjoc.14.109 |
| 21. | Figueras, E.; Borbély, A.; Ismail, M.; Frese, M.; Sewald, N. Beilstein J. Org. Chem. 2018, 14, 1281–1286. doi:10.3762/bjoc.14.109 |
| 2. | Schwartz, R. E.; Hirsch, C. F.; Sesin, D. F.; Flor, J. E.; Chartrain, M.; Fromtling, R. E.; Harris, G. H.; Salvatore, M. J.; Liesch, J. M.; Yudin, K. J. Ind. Microbiol. 1990, 5, 113–123. doi:10.1007/bf01573860 |
| 20. | Patel, V. F.; Andis, S. L.; Kennedy, J. H.; Ray, J. E.; Schultz, R. M. J. Med. Chem. 1999, 42, 2588–2603. doi:10.1021/jm980706s |
| 20. | Patel, V. F.; Andis, S. L.; Kennedy, J. H.; Ray, J. E.; Schultz, R. M. J. Med. Chem. 1999, 42, 2588–2603. doi:10.1021/jm980706s |
| 5. | Lai, Q.; Wu, M.; Wang, R.; Lai, W.; Tao, Y.; Lu, Y.; Wang, Y.; Yu, L.; Zhang, R.; Peng, Y.; Jiang, X.; Fu, Y.; Wang, X.; Zhang, Z.; Guo, C.; Liao, W.; Zhang, Y.; Kang, T.; Chen, H.; Yao, Y.; Gou, L.; Yang, J. Eur. J. Med. Chem. 2020, 199, 112364. doi:10.1016/j.ejmech.2020.112364 |
| 12. | Figueras, E.; Martins, A.; Borbély, A.; Le Joncour, V.; Cordella, P.; Perego, R.; Modena, D.; Pagani, P.; Esposito, S.; Auciello, G.; Frese, M.; Gallinari, P.; Laakkonen, P.; Steinkühler, C.; Sewald, N. Pharmaceutics 2019, 11, 220. doi:10.3390/pharmaceutics11050220 |
| 13. | Anselmi, M.; Borbély, A.; Figueras, E.; Michalek, C.; Kemker, I.; Gentilucci, L.; Sewald, N. Chem. – Eur. J. 2021, 27, 1015–1022. doi:10.1002/chem.202003471 |
| 14. | Georg, G. I.; Ali, S. M.; Stella, V. J.; Waugh, W. N.; Himes, R. H. Bioorg. Med. Chem. Lett. 1998, 8, 1959–1962. doi:10.1016/s0960-894x(98)00356-4 |
| 15. | Cazzamalli, S.; Figueras, E.; Pethő, L.; Borbély, A.; Steinkühler, C.; Neri, D.; Sewald, N. ACS Omega 2018, 3, 14726–14731. doi:10.1021/acsomega.8b02350 |
| 16. | Borbély, A.; Figueras, E.; Martins, A.; Esposito, S.; Auciello, G.; Monteagudo, E.; Di Marco, A.; Summa, V.; Cordella, P.; Perego, R.; Kemker, I.; Frese, M.; Gallinari, P.; Steinkühler, C.; Sewald, N. Pharmaceutics 2019, 11, 151. doi:10.3390/pharmaceutics11040151 |
| 17. | Borbély, A.; Figueras, E.; Martins, A.; Bodero, L.; Raposo Moreira Dias, A.; López Rivas, P.; Pina, A.; Arosio, D.; Gallinari, P.; Frese, M.; Steinkühler, C.; Gennari, C.; Piarulli, U.; Sewald, N. ChemistryOpen 2019, 8, 737–742. doi:10.1002/open.201900110 |
| 19. | Dessin, C.; Schachtsiek, T.; Voss, J.; Abel, A.-C.; Neumann, B.; Stammler, H.-G.; Prota, A. E.; Sewald, N. Angew. Chem., Int. Ed. 2024, 63, e202416210. doi:10.1002/anie.202416210 |
| 20. | Patel, V. F.; Andis, S. L.; Kennedy, J. H.; Ray, J. E.; Schultz, R. M. J. Med. Chem. 1999, 42, 2588–2603. doi:10.1021/jm980706s |
| 35. | Although the tissue type of the KB-3-1 cell line (cervical carcinoma) and the CCRF-CEM cell line (T lymphoblasts) are different, there is generally only a very small difference in the IC50 values for many different cryptophycins (see Sewald et al., [8]), so that we assume good comparability of the IC50 values between the two cell lines. |
| 9. | Bouchard, H.; Brun, M.-P.; Hubert, P. Novel peptidic linkers and cryptophycin conjugates, their preparation and their therapeutic use. U.S. Pat. Appl. US20210228726A1, July 29, 2021. |
| 10. | McGonigle, S.; Majumder, U.; Custar, D. W.; Wu, J.; Postema, M. H. D. Peptide drug conjugates. WO Pat. Appl. WO2017136769A1, Aug 10, 2017. |
| 11. | Eißler, S.; Bogner, T.; Nahrwold, M.; Sewald, N. Chem. – Eur. J. 2009, 15, 11273–11287. doi:10.1002/chem.200901750 |
| 20. | Patel, V. F.; Andis, S. L.; Kennedy, J. H.; Ray, J. E.; Schultz, R. M. J. Med. Chem. 1999, 42, 2588–2603. doi:10.1021/jm980706s |
| 19. | Dessin, C.; Schachtsiek, T.; Voss, J.; Abel, A.-C.; Neumann, B.; Stammler, H.-G.; Prota, A. E.; Sewald, N. Angew. Chem., Int. Ed. 2024, 63, e202416210. doi:10.1002/anie.202416210 |
| 8. | Weiss, C.; Figueras, E.; Borbely, A. N.; Sewald, N. J. Pept. Sci. 2017, 23, 514–531. doi:10.1002/psc.3015 |
| 19. | Dessin, C.; Schachtsiek, T.; Voss, J.; Abel, A.-C.; Neumann, B.; Stammler, H.-G.; Prota, A. E.; Sewald, N. Angew. Chem., Int. Ed. 2024, 63, e202416210. doi:10.1002/anie.202416210 |
| 7. | McCombs, J. R.; Owen, S. C. AAPS J. 2015, 17, 339–351. doi:10.1208/s12248-014-9710-8 |
| 18. | Nahrwold, M.; Weiß, C.; Bogner, T.; Mertink, F.; Conradi, J.; Sammet, B.; Palmisano, R.; Royo Gracia, S.; Preuße, T.; Sewald, N. J. Med. Chem. 2013, 56, 1853–1864. doi:10.1021/jm301346z |
| 33. | Akiyama, S.-i.; Fojo, A.; Hanover, J. A.; Pastan, I.; Gottesman, M. M. Somatic Cell Mol. Genet. 1985, 11, 117–126. doi:10.1007/bf01534700 |
| 34. | Shen, D. W.; Cardarelli, C.; Hwang, J.; Cornwell, M.; Richert, N.; Ishii, S.; Pastan, I.; Gottesman, M. M. J. Biol. Chem. 1986, 261, 7762–7770. doi:10.1016/s0021-9258(19)57466-x |
| 23. | Wang, L.; Hisano, W.; Murai, Y.; Sakurai, M.; Muto, Y.; Ikemoto, H.; Okamoto, M.; Murotani, T.; Isoda, R.; Kim, D.; Sakihama, Y.; Sitepu, I. R.; Hashidoko, Y.; Hatanaka, Y.; Hashimoto, M. Molecules 2013, 18, 8393–8401. doi:10.3390/molecules18078393 |
| 20. | Patel, V. F.; Andis, S. L.; Kennedy, J. H.; Ray, J. E.; Schultz, R. M. J. Med. Chem. 1999, 42, 2588–2603. doi:10.1021/jm980706s |
| 8. | Weiss, C.; Figueras, E.; Borbely, A. N.; Sewald, N. J. Pept. Sci. 2017, 23, 514–531. doi:10.1002/psc.3015 |
| 21. | Figueras, E.; Borbély, A.; Ismail, M.; Frese, M.; Sewald, N. Beilstein J. Org. Chem. 2018, 14, 1281–1286. doi:10.3762/bjoc.14.109 |
| 28. | Eissler, S.; Nahrwold, M.; Neumann, B.; Stammler, H.-G.; Sewald, N. Org. Lett. 2007, 9, 817–819. doi:10.1021/ol063032l |
| 29. | Sammet, B.; Bogner, T.; Nahrwold, M.; Weiss, C.; Sewald, N. J. Org. Chem. 2010, 75, 6953–6960. doi:10.1021/jo101563s |
| 30. | Nahrwold, M.; Bogner, T.; Eissler, S.; Verma, S.; Sewald, N. Org. Lett. 2010, 12, 1064–1067. doi:10.1021/ol1000473 |
| 11. | Eißler, S.; Bogner, T.; Nahrwold, M.; Sewald, N. Chem. – Eur. J. 2009, 15, 11273–11287. doi:10.1002/chem.200901750 |
| 26. | Tripathy, N. K.; Georg, G. I. Tetrahedron Lett. 2004, 45, 5309–5311. doi:10.1016/j.tetlet.2004.04.164 |
| 27. | Bendelac, A.; Benedetti, F.; Doublet-Decabras, V.; Lokovi, R.; Decalogne, F.; Bigot, A. Org. Process Res. Dev. 2022, 26, 2145–2154. doi:10.1021/acs.oprd.2c00080 |
| 21. | Figueras, E.; Borbély, A.; Ismail, M.; Frese, M.; Sewald, N. Beilstein J. Org. Chem. 2018, 14, 1281–1286. doi:10.3762/bjoc.14.109 |
| 20. | Patel, V. F.; Andis, S. L.; Kennedy, J. H.; Ray, J. E.; Schultz, R. M. J. Med. Chem. 1999, 42, 2588–2603. doi:10.1021/jm980706s |
| 21. | Figueras, E.; Borbély, A.; Ismail, M.; Frese, M.; Sewald, N. Beilstein J. Org. Chem. 2018, 14, 1281–1286. doi:10.3762/bjoc.14.109 |
| 24. | Arns, S. P.; Jaquith, J. B.; Baudelet, D. J.; Simonson, E. R.; Liggins, R. T.; Kumar, N. S.; Hsieh, T. H. H. Somatostatin receptor antagonist compounds and methods of using the same. WO Patent WO2017/136943, Aug 17, 2017. |
| 25. | Byun, E.; Hong, B.; De Castro, K. A.; Lim, M.; Rhee, H. J. Org. Chem. 2007, 72, 9815–9817. doi:10.1021/jo701503q |
© 2025 Schachtsiek et al.; licensee Beilstein-Institut.
This is an open access article licensed under the terms of the Beilstein-Institut Open Access License Agreement (https://www.beilstein-journals.org/bjoc/terms), which is identical to the Creative Commons Attribution 4.0 International License (https://creativecommons.org/licenses/by/4.0). The reuse of material under this license requires that the author(s), source and license are credited. Third-party material in this article could be subject to other licenses (typically indicated in the credit line), and in this case, users are required to obtain permission from the license holder to reuse the material.